Gene Page: MYD88
Summary ?
| GeneID | 4615 |
| Symbol | MYD88 |
| Synonyms | MYD88D |
| Description | myeloid differentiation primary response 88 |
| Reference | MIM:602170|HGNC:HGNC:7562|Ensembl:ENSG00000172936|HPRD:03703|Vega:OTTHUMG00000131083 |
| Gene type | protein-coding |
| Map location | 3p22 |
| Pascal p-value | 0.205 |
| Fetal beta | 2.387 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2070396 | chr21 | 30878751 | MYD88 | 4615 | 0.12 | trans |
Section II. Transcriptome annotation
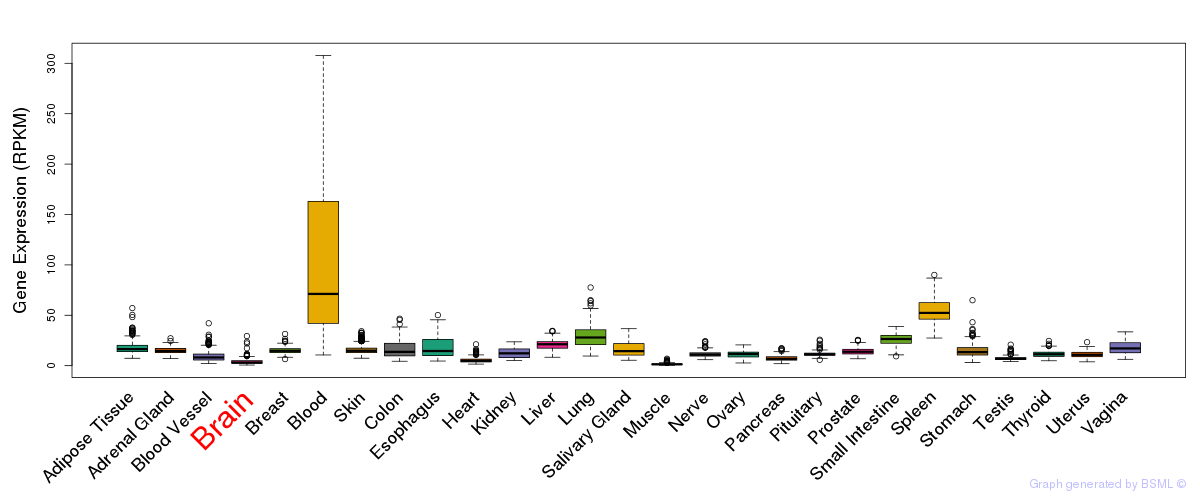
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SDC3 | 0.73 | 0.69 |
| SLC7A5 | 0.72 | 0.69 |
| TBC1D2B | 0.71 | 0.73 |
| RAB3IL1 | 0.70 | 0.71 |
| DENND2A | 0.69 | 0.66 |
| ABLIM1 | 0.67 | 0.63 |
| ITGA3 | 0.67 | 0.61 |
| PCDHGB8P | 0.66 | 0.65 |
| KIAA1539 | 0.66 | 0.61 |
| CUEDC1 | 0.66 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCRG1 | -0.53 | -0.40 |
| AL050337.1 | -0.51 | -0.42 |
| COX7B | -0.51 | -0.49 |
| SYCP3 | -0.50 | -0.42 |
| MT3 | -0.49 | -0.36 |
| AL138743.2 | -0.49 | -0.37 |
| C1orf54 | -0.49 | -0.42 |
| UQCRB | -0.49 | -0.48 |
| MYL3 | -0.48 | -0.40 |
| LY96 | -0.48 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | IEA | - | |
| GO:0005123 | death receptor binding | TAS | 9374458 | |
| GO:0005515 | protein binding | IPI | 10383454 |10880445 |11544529 |16286016 |16365431 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | IEA | - | |
| GO:0002238 | response to molecule of fungal origin | IEA | - | |
| GO:0016064 | immunoglobulin mediated immune response | IEA | - | |
| GO:0009615 | response to virus | IEA | - | |
| GO:0006954 | inflammatory response | IEA | - | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IEA | - | |
| GO:0032496 | response to lipopolysaccharide | IEA | - | |
| GO:0032760 | positive regulation of tumor necrosis factor production | IEA | - | |
| GO:0031663 | lipopolysaccharide-mediated signaling pathway | IEA | - | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEP | 12761501 | |
| GO:0045087 | innate immune response | IEA | - | |
| GO:0045080 | positive regulation of chemokine biosynthetic process | IEA | - | |
| GO:0045351 | type I interferon biosynthetic process | IEA | - | |
| GO:0046330 | positive regulation of JNK cascade | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0031315 | extrinsic to mitochondrial outer membrane | IEA | - | |
| GO:0031224 | intrinsic to membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 11828002 |
| IL1R1 | CD121A | D2S1473 | IL-1R-alpha | IL1R | IL1RA | P80 | interleukin 1 receptor, type I | Affinity Capture-Western | BioGRID | 10854325 |
| IL1RAP | C3orf13 | FLJ37788 | IL-1RAcP | IL1R3 | interleukin 1 receptor accessory protein | - | HPRD,BioGRID | 11880380 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | IRAK does not interact with MyD88, whilst the kinase inactive mutant IRAK[Lys239Ser] does interact. | BIND | 10383454 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | - | HPRD | 10854325 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | Affinity Capture-Western | BioGRID | 10383454 |11976320 |
| IRAK2 | IRAK-2 | MGC150550 | interleukin-1 receptor-associated kinase 2 | IRAK-2 interacts with MyD88. | BIND | 10383454 |
| IRAK2 | IRAK-2 | MGC150550 | interleukin-1 receptor-associated kinase 2 | - | HPRD,BioGRID | 9374458 |
| IRAK3 | ASRT5 | IRAK-M | IRAKM | interleukin-1 receptor-associated kinase 3 | IRAK-M interacts with MyD88. | BIND | 10383454 |
| IRAK3 | ASRT5 | IRAK-M | IRAKM | interleukin-1 receptor-associated kinase 3 | Affinity Capture-Western | BioGRID | 10383454 |
| IRAK4 | IPD1 | NY-REN-64 | REN64 | interleukin-1 receptor-associated kinase 4 | - | HPRD,BioGRID | 11960013 |
| IRF7 | IRF-7H | IRF7A | interferon regulatory factor 7 | IRF7 interacts with MyD88. | BIND | 15361868 |
| PELI2 | - | pellino homolog 2 (Drosophila) | - | HPRD | 12860405 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11416133 |
| SPOP | TEF2 | speckle-type POZ protein | Two-hybrid | BioGRID | 16189514 |
| TIRAP | FLJ42305 | Mal | wyatt | toll-interleukin 1 receptor (TIR) domain containing adaptor protein | - | HPRD,BioGRID | 11544529 |
| TLR2 | CD282 | TIL4 | toll-like receptor 2 | Two-hybrid | BioGRID | 15107846 |
| TLR3 | CD283 | toll-like receptor 3 | - | HPRD,BioGRID | 12646618 |
| TLR4 | ARMD10 | CD284 | TOLL | hToll | toll-like receptor 4 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10952994 |12646618 |15107846 |
| TLR4 | ARMD10 | CD284 | TOLL | hToll | toll-like receptor 4 | - | HPRD | 10952994 |11743586 |
| TLR5 | FLJ10052 | MGC126430 | MGC126431 | SLEB1 | TIL3 | toll-like receptor 5 | Two-hybrid | BioGRID | 15107846 |
| TLR9 | CD289 | toll-like receptor 9 | Two-hybrid | BioGRID | 15107846 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TOLL ENDOGENOUS PATHWAY | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR SIGNALS VIA NFKB | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP | 52 | 38 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C3 | 20 | 15 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN | 108 | 64 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8 | 49 | 36 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| VANLOO SP3 TARGETS DN | 89 | 47 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |