Gene Page: NFATC2
Summary ?
| GeneID | 4773 |
| Symbol | NFATC2 |
| Synonyms | NFAT1|NFATP |
| Description | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| Reference | MIM:600490|HGNC:HGNC:7776|Ensembl:ENSG00000101096|HPRD:02730|Vega:OTTHUMG00000032747 |
| Gene type | protein-coding |
| Map location | 20q13.2 |
| Pascal p-value | 0.714 |
| Fetal beta | -0.478 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0218 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
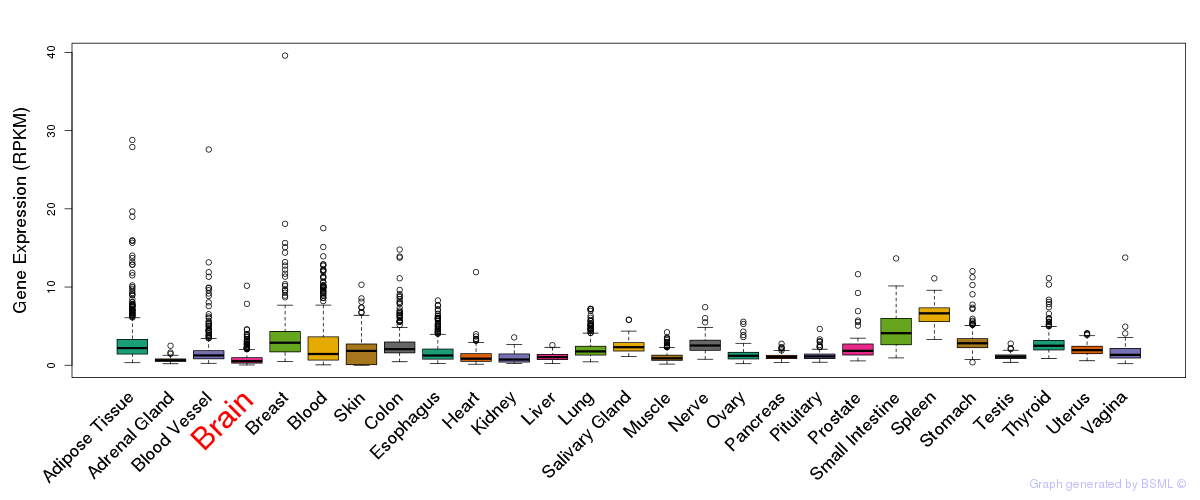
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC015656.1 | 0.91 | 0.81 |
| GPR139 | 0.68 | 0.73 |
| KLF11 | 0.67 | 0.72 |
| LRRC55 | 0.66 | 0.75 |
| FAM129B | 0.66 | 0.75 |
| EFNB2 | 0.66 | 0.72 |
| AC023424.3 | 0.65 | 0.71 |
| AC010974.2 | 0.65 | 0.76 |
| LCMT2 | 0.65 | 0.74 |
| AC010265.1 | 0.65 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.44 | -0.71 |
| CA4 | -0.40 | -0.58 |
| PTH1R | -0.40 | -0.54 |
| AF347015.31 | -0.40 | -0.71 |
| ACOT13 | -0.39 | -0.60 |
| AIFM3 | -0.39 | -0.56 |
| RGN | -0.39 | -0.53 |
| CMTM8 | -0.39 | -0.49 |
| MT-CO2 | -0.39 | -0.70 |
| TNFSF12 | -0.39 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 8668213 | |
| GO:0005515 | protein binding | IPI | 16873067 | |
| GO:0016563 | transcription activator activity | IDA | 15790681 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001816 | cytokine production | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | TAS | 8668213 | |
| GO:0045941 | positive regulation of transcription | IDA | 15790681 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 8668213 | |
| GO:0005737 | cytoplasm | TAS | 8668213 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 9625762 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | - | HPRD | 12560487 |
| EGR4 | NGFI-C | NGFIC | PAT133 | early growth response 4 | - | HPRD | 12560487 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 9625762 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 8397339 |12949493 |
| IRF4 | LSIRF | MUM1 | interferon regulatory factor 4 | - | HPRD,BioGRID | 11956291 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | - | HPRD,BioGRID | 10944115 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 12949493 |
| NFATC2IP | FLJ14639 | MGC126790 | MGC138387 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein | - | HPRD | 8943202 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD,BioGRID | 11356192 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 12021260 |
| SNRK | DKFZp779A1866 | FLJ20224 | HSNFRK | KIAA0096 | SNF related kinase | - | HPRD | 10878360 |12087103 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | - | HPRD | 10821850 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD,BioGRID | 10611249 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 10611249 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CALCINEURIN PATHWAY | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GATA3 PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE UP | 136 | 80 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION APOBEC2 | 27 | 19 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| CHEMELLO SOLEUS VS EDL MYOFIBERS UP | 35 | 16 | All SZGR 2.0 genes in this pathway |