Gene Page: NFKB2
Summary ?
| GeneID | 4791 |
| Symbol | NFKB2 |
| Synonyms | CVID10|H2TF1|LYT-10|LYT10|NF-kB2|p100|p49/p100|p52 |
| Description | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 |
| Reference | MIM:164012|HGNC:HGNC:7795|Ensembl:ENSG00000077150|HPRD:01239|Vega:OTTHUMG00000018962 |
| Gene type | protein-coding |
| Map location | 10q24 |
| Pascal p-value | 0.005 |
| Fetal beta | -0.659 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0233 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23738770 | 10 | 104154605 | NFKB2 | -0.025 | 0.3 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
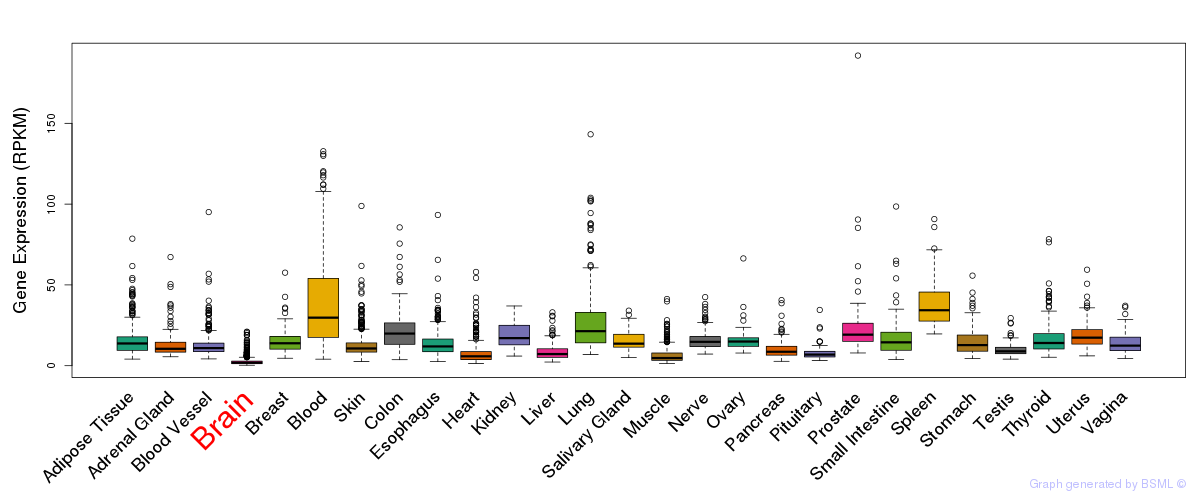
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 1760839 |1876189 | |
| GO:0003713 | transcription coactivator activity | TAS | 1876189 | |
| GO:0005515 | protein binding | IPI | 11526476 |16108830 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002268 | follicular dendritic cell differentiation | IEA | dendrite (GO term level: 5) | - |
| GO:0002467 | germinal center formation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IDA | 8360178 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0030198 | extracellular matrix organization | IEA | - | |
| GO:0048536 | spleen development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10723127 | |
| GO:0005634 | nucleus | IDA | 15677444 | |
| GO:0005654 | nucleoplasm | EXP | 10723127 | |
| GO:0005737 | cytoplasm | IDA | 15677444 | |
| GO:0033257 | Bcl3/NF-kappaB2 complex | IDA | 9407099 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AIFM1 | AIF | MGC111425 | PDCD8 | apoptosis-inducing factor, mitochondrion-associated, 1 | - | HPRD | 14743216 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | p52 interacts with BCL-3. This interaction was modelled on a demonstrated interaction between p52 from an unspecified species and human BCL-3. | BIND | 15469820 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD | 8453667 |11707390 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | Co-localization Reconstituted Complex | BioGRID | 8453667 |14678988 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | - | HPRD,BioGRID | 11994270 |
| DDX3X | DBX | DDX14 | DDX3 | HLP2 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | Affinity Capture-MS | BioGRID | 14743216 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD | 14743216 |
| DSP | DPI | DPII | desmoplakin | - | HPRD | 14743216 |
| FBXW7 | AGO | CDC4 | DKFZp686F23254 | FBW6 | FBW7 | FBX30 | FBXO30 | FBXW6 | FLJ16457 | SEL-10 | SEL10 | F-box and WD repeat domain containing 7 | Affinity Capture-MS | BioGRID | 14743216 |
| GLG1 | CFR-1 | ESL-1 | FLJ23319 | FLJ23967 | MG-160 | MG160 | golgi apparatus protein 1 | Affinity Capture-MS | BioGRID | 14743216 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD | 14743216 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | - | HPRD | 14743216 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | Affinity Capture-MS | BioGRID | 14743216 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11239468 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | Affinity Capture-MS | BioGRID | 14743216 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | - | HPRD,BioGRID | 11526476 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | Affinity Capture-MS | BioGRID | 14743216 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | Affinity Capture-MS | BioGRID | 14743216 |
| NFKBIE | IKBE | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | p52 interacts with I-kappa-B-epsilon. This interaction was modeled on a demonstrated interaction between p52 from an unspecified species and human I-kappa-B-epsilon. | BIND | 9135156 |
| NFKBIE | IKBE | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | - | HPRD,BioGRID | 9315679 |
| NKRF | ITBA4 | NRF | NFKB repressing factor | - | HPRD,BioGRID | 10562553 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 7823959 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | - | HPRD | 14743216 |
| PSMD1 | MGC133040 | MGC133041 | P112 | Rpn2 | S1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | - | HPRD | 14743216 |
| PSMD3 | P58 | RPN3 | S3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | - | HPRD | 14743216 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 8413211 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 8413211 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | Affinity Capture-MS Co-localization | BioGRID | 14678988 |14743216 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | - | HPRD | 12505990 |
| RPL18 | - | ribosomal protein L18 | - | HPRD | 14743216 |
| RPL30 | - | ribosomal protein L30 | Affinity Capture-MS | BioGRID | 14743216 |
| RPL6 | SHUJUN-2 | TAXREB107 | TXREB1 | ribosomal protein L6 | Affinity Capture-MS | BioGRID | 14743216 |
| RPS13 | - | ribosomal protein S13 | Affinity Capture-MS | BioGRID | 14743216 |
| RPS3 | FLJ26283 | FLJ27450 | MGC87870 | ribosomal protein S3 | - | HPRD | 14743216 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | - | HPRD | 14743216 |
| SEC16A | FLJ26737 | KIAA0310 | RP11-413M3.10 | SEC16L | p250 | SEC16 homolog A (S. cerevisiae) | - | HPRD,BioGRID | 14743216 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 7933095 |
| TBK1 | FLJ11330 | NAK | T2K | TANK-binding kinase 1 | Affinity Capture-MS | BioGRID | 14743216 |
| TIMM50 | MGC102733 | TIM50 | TIM50L | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | - | HPRD | 14743216 |
| TNIP2 | ABIN-2 | ABIN2 | FLIP1 | KLIP | MGC4289 | TNFAIP3 interacting protein 2 | - | HPRD,BioGRID | 14743216 |
| TSC22D3 | DIP | DKFZp313A1123 | DSIPI | GILZ | TSC-22R | hDIP | TSC22 domain family, member 3 | - | HPRD,BioGRID | 11468175 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | - | HPRD | 14743216 |
| TUBB6 | HsT1601 | MGC132410 | MGC4083 | TUBB-5 | tubulin, beta 6 | - | HPRD | 14743216 |
| TUBGCP2 | GCP2 | MGC138162 | SPBC97 | Spc97p | tubulin, gamma complex associated protein 2 | - | HPRD | 14743216 |
| USP2 | UBP41 | USP9 | ubiquitin specific peptidase 2 | Affinity Capture-MS | BioGRID | 14743216 |
| USP9X | DFFRX | FAF | FAM | ubiquitin specific peptidase 9, X-linked | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST GAQ PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| GILMORE CORE NFKB PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK TARGETS | 18 | 15 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| RASHI NFKB1 TARGETS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 3 | 28 | 23 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS TEMPORAL UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO CURCUMIN SULINDAC 7 | 19 | 10 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION DN | 13 | 7 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING VIA NFKB | 28 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| PHONG TNF TARGETS UP | 63 | 43 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |