Gene Page: NGFR
Summary ?
| GeneID | 4804 |
| Symbol | NGFR |
| Synonyms | CD271|Gp80-LNGFR|TNFRSF16|p75(NTR)|p75NTR |
| Description | nerve growth factor receptor |
| Reference | MIM:162010|HGNC:HGNC:7809|Ensembl:ENSG00000064300|HPRD:01197|Vega:OTTHUMG00000161495 |
| Gene type | protein-coding |
| Map location | 17q21-q22 |
| Pascal p-value | 0.328 |
| Fetal beta | 0.618 |
| eGene | Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9908695 | 17 | 47550976 | NGFR | ENSG00000064300.4 | 2.01019E-6 | 0.01 | -21679 | gtex_brain_putamen_basal |
| rs1317636 | 17 | 47551127 | NGFR | ENSG00000064300.4 | 4.19031E-7 | 0.01 | -21528 | gtex_brain_putamen_basal |
| rs871882 | 17 | 47552043 | NGFR | ENSG00000064300.4 | 4.66135E-7 | 0.01 | -20612 | gtex_brain_putamen_basal |
| rs62077745 | 17 | 47552169 | NGFR | ENSG00000064300.4 | 1.65035E-6 | 0.01 | -20486 | gtex_brain_putamen_basal |
| rs62077746 | 17 | 47553947 | NGFR | ENSG00000064300.4 | 2.12322E-6 | 0.01 | -18708 | gtex_brain_putamen_basal |
| rs62077747 | 17 | 47555501 | NGFR | ENSG00000064300.4 | 2.24691E-6 | 0.01 | -17154 | gtex_brain_putamen_basal |
| rs58700210 | 17 | 47556197 | NGFR | ENSG00000064300.4 | 2.30671E-6 | 0.01 | -16458 | gtex_brain_putamen_basal |
| rs2001868 | 17 | 47559207 | NGFR | ENSG00000064300.4 | 1.26252E-6 | 0.01 | -13448 | gtex_brain_putamen_basal |
| rs2584667 | 17 | 47560071 | NGFR | ENSG00000064300.4 | 1.22511E-6 | 0.01 | -12584 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
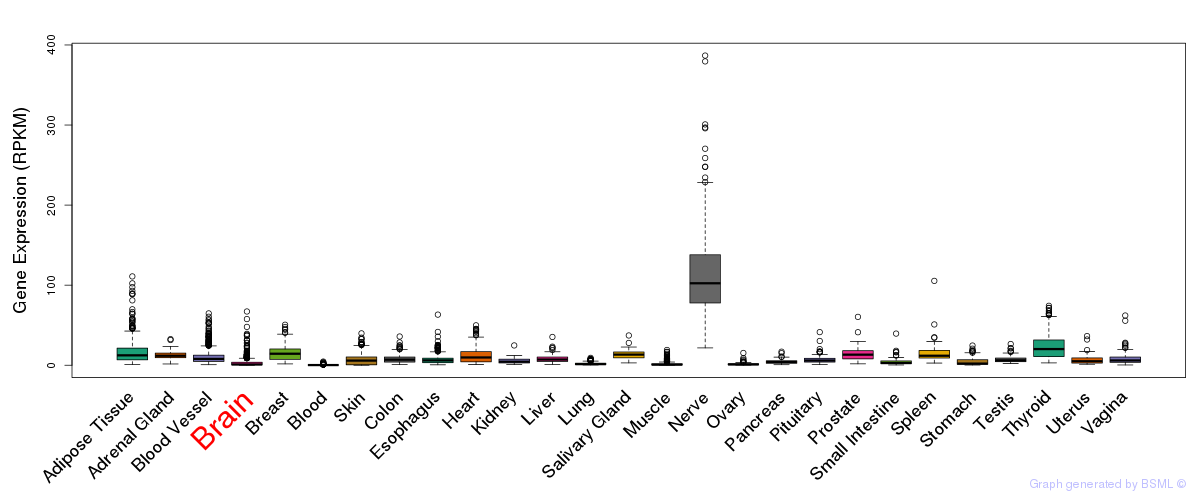
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPS25 | 0.85 | 0.85 |
| C17orf79 | 0.85 | 0.87 |
| TPI1 | 0.84 | 0.86 |
| MECR | 0.84 | 0.85 |
| SNRPN | 0.83 | 0.85 |
| NDUFA8 | 0.83 | 0.85 |
| SNX17 | 0.82 | 0.84 |
| STUB1 | 0.82 | 0.84 |
| TMEM111 | 0.82 | 0.84 |
| DHRS7B | 0.82 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAP4K4 | -0.44 | -0.41 |
| THOC2 | -0.43 | -0.40 |
| TTC28 | -0.43 | -0.28 |
| ATAD2B | -0.43 | -0.35 |
| AC005035.1 | -0.43 | -0.41 |
| GJC1 | -0.42 | -0.36 |
| EIF2C4 | -0.42 | -0.25 |
| SRGAP1 | -0.42 | -0.27 |
| NHSL1 | -0.42 | -0.37 |
| MBTD1 | -0.42 | -0.25 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005035 | death receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0048406 | nerve growth factor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEA | Brain (GO term level: 6) | - |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0016048 | detection of temperature stimulus | IEA | - | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0021675 | nerve development | IEA | - | |
| GO:0042488 | positive regulation of odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0043588 | skin development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | TAS | 3022937 | |
| GO:0005887 | integral to plasma membrane | TAS | 3022937 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | Reconstituted Complex | BioGRID | 9867838 |
| FSCN1 | FLJ38511 | SNL | p55 | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12789270 |
| KIDINS220 | ARMS | MGC163482 | kinase D-interacting substrate, 220kDa | - | HPRD | 11150334 |15378608 |
| LINGO1 | FLJ14594 | LERN1 | LRRN6A | MGC17422 | UNQ201 | leucine rich repeat and Ig domain containing 1 | LINGO-1 interacts with p75. This interaction was modeled on a demonstrated interaction between LINGO-1 from human and p75 from rat. | BIND | 14966521 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD | 12011108 |
| MAGED1 | DLXIN-1 | NRAGE | melanoma antigen family D, 1 | Affinity Capture-Western | BioGRID | 12716928 |
| MAGEH1 | APR-1 | APR1 | MAGEH | melanoma antigen family H, 1 | Affinity Capture-Western | BioGRID | 12414813 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | - | HPRD,BioGRID | 12414813 |12716928 |14593116 |
| NDNL2 | HCA4 | MAGEG1 | MAGEL3 | NSE3 | NSMCE3 | necdin-like 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14593116 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | - | HPRD,BioGRID | 14985763 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | NGF interacts with p75NTR. | BIND | 14985763 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | p75NTR interacts with NGF. | BIND | 15258592 |
| NGFRAP1 | BEX3 | Bex | DXS6984E | HGR74 | NADE | nerve growth factor receptor (TNFRSF16) associated protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10764727 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | p75NTR interacts with NT-4. | BIND | 15258592 |
| PRKACB | DKFZp781I2452 | MGC41879 | MGC9320 | PKACB | protein kinase, cAMP-dependent, catalytic, beta | Affinity Capture-Western Biochemical Activity Two-hybrid | BioGRID | 12682012 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | p75 interacts with NgR. This interaction was modeled on a demonstrated interaction between p75 and NgR both from an unspecified species. | BIND | 15953414 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | p75 interacts with NgR. This interaction was modelled on a demonstrated interaction between rat p75 and human NgR. | BIND | 15694321 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | - | HPRD | 12422217 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | p75 interacts with NgR1. This interaction was modeled on a demonstrated interaction between p75 from rat and NgR1 from human. | BIND | 14966521 |
| SFRS10 | DKFZp686F18120 | Htra2-beta | SRFS10 | TRA2-BETA | TRA2B | TRAN2B | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | - | HPRD | 12604596 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 7541035 |
| SORT1 | Gp95 | NT3 | sortilin 1 | p75NTR interacts with sortilin. This interaction was modeled on a demonstrated interaction between p75NTR from human and sortilin from an unspecified species. | BIND | 14985763 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | Affinity Capture-Western | BioGRID | 12604596 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | TRADD interacts with NGFR (p75NTR). | BIND | 12604596 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | Affinity Capture-Western | BioGRID | 10514511 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 10514511 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | Affinity Capture-Western | BioGRID | 10514511 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | - | HPRD,BioGRID | 10514511 |
| TRAF5 | MGC:39780 | RNF84 | TNF receptor-associated factor 5 | Affinity Capture-Western | BioGRID | 10514511 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10514511 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NGF PATHWAY | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDK5 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| SA TRKA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATED PROTEOLYSIS OF P75NTR | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME P75NTR SIGNALS VIA NFKB | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI RESPONSE TO ROMIDEPSIN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY DEACETYLATION IN PANCREATIC CANCER | 50 | 30 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 30 | 36 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-128 | 429 | 436 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-143 | 261 | 267 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-27 | 430 | 437 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-299-3p | 27 | 33 | m8 | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.