Gene Page: NOV
Summary ?
| GeneID | 4856 |
| Symbol | NOV |
| Synonyms | CCN3|IBP-9|IGFBP-9|IGFBP9|NOVh |
| Description | nephroblastoma overexpressed |
| Reference | MIM:164958|HGNC:HGNC:7885|Ensembl:ENSG00000136999|HPRD:01291|Vega:OTTHUMG00000164984 |
| Gene type | protein-coding |
| Map location | 8q24.1 |
| Pascal p-value | 0.653 |
| Fetal beta | -0.95 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17535450 | chr2 | 39900012 | NOV | 4856 | 0.01 | trans | ||
| rs11706314 | chr3 | 45309716 | NOV | 4856 | 3.461E-5 | trans | ||
| rs11706394 | chr3 | 45309907 | NOV | 4856 | 0 | trans | ||
| rs2922180 | chr3 | 125280472 | NOV | 4856 | 0.05 | trans | ||
| rs2976809 | chr3 | 125451738 | NOV | 4856 | 0.02 | trans | ||
| rs17090446 | chr4 | 62255221 | NOV | 4856 | 0.03 | trans | ||
| rs7668550 | chr4 | 104384355 | NOV | 4856 | 0.07 | trans | ||
| rs17034860 | chr4 | 135177933 | NOV | 4856 | 0.13 | trans | ||
| rs16894149 | chr5 | 61013724 | NOV | 4856 | 0.02 | trans | ||
| rs6449550 | chr5 | 61034312 | NOV | 4856 | 0.01 | trans | ||
| rs4489051 | chr5 | 147954484 | NOV | 4856 | 0.04 | trans | ||
| rs17056435 | chr5 | 158453782 | NOV | 4856 | 0.02 | trans | ||
| rs9313797 | chr5 | 158465457 | NOV | 4856 | 0.01 | trans | ||
| rs17056474 | chr5 | 158468105 | NOV | 4856 | 0.01 | trans | ||
| rs2294262 | chr6 | 16108966 | NOV | 4856 | 0.06 | trans | ||
| rs9321275 | chr6 | 131493217 | NOV | 4856 | 0.07 | trans | ||
| rs6593322 | chr7 | 51048058 | NOV | 4856 | 0.17 | trans | ||
| rs2199402 | chr8 | 9201002 | NOV | 4856 | 0.04 | trans | ||
| rs7841407 | chr8 | 9243427 | NOV | 4856 | 0 | trans | ||
| rs9325776 | chr8 | 15817555 | NOV | 4856 | 0.19 | trans | ||
| rs16929424 | chr8 | 63608389 | NOV | 4856 | 0.15 | trans | ||
| rs16908622 | chr8 | 139231604 | NOV | 4856 | 0.06 | trans | ||
| rs2066720 | chr9 | 107554068 | NOV | 4856 | 0.02 | trans | ||
| rs9423711 | chr10 | 2574802 | NOV | 4856 | 0.04 | trans | ||
| rs4514358 | chr10 | 43861923 | NOV | 4856 | 0.01 | trans | ||
| rs822083 | chr10 | 108713231 | NOV | 4856 | 0.11 | trans | ||
| rs11820030 | chr11 | 76133564 | NOV | 4856 | 0 | trans | ||
| rs17134872 | chr11 | 76139845 | NOV | 4856 | 2.289E-4 | trans | ||
| rs10751255 | 0 | NOV | 4856 | 0.02 | trans | |||
| rs11236774 | chr11 | 76234952 | NOV | 4856 | 2.289E-4 | trans | ||
| rs9532792 | chr13 | 41912654 | NOV | 4856 | 0.05 | trans | ||
| rs9590586 | chr13 | 41913130 | NOV | 4856 | 0.02 | trans | ||
| rs7358856 | chr13 | 41931609 | NOV | 4856 | 0.02 | trans | ||
| rs7358853 | chr13 | 41931685 | NOV | 4856 | 0.01 | trans | ||
| rs8002343 | chr13 | 41952090 | NOV | 4856 | 0.02 | trans | ||
| rs9315839 | chr13 | 41952872 | NOV | 4856 | 0.04 | trans | ||
| rs4349050 | chr13 | 41958722 | NOV | 4856 | 0 | trans | ||
| rs7982172 | chr13 | 41969697 | NOV | 4856 | 7.174E-4 | trans | ||
| rs4942043 | chr13 | 41978218 | NOV | 4856 | 9.529E-4 | trans | ||
| rs4402447 | chr13 | 41984741 | NOV | 4856 | 0 | trans | ||
| rs1750009 | chr13 | 42657093 | NOV | 4856 | 0.09 | trans | ||
| rs9529974 | chr13 | 72821935 | NOV | 4856 | 0.06 | trans | ||
| rs17720221 | chr13 | 75182377 | NOV | 4856 | 0.05 | trans | ||
| rs10149864 | chr14 | 86719683 | NOV | 4856 | 0.15 | trans | ||
| rs2093759 | chr14 | 97171024 | NOV | 4856 | 0.13 | trans | ||
| rs17244419 | chr14 | 97171074 | NOV | 4856 | 0.06 | trans | ||
| rs11073814 | chr15 | 89378883 | NOV | 4856 | 0.1 | trans | ||
| rs7165622 | chr15 | 93979910 | NOV | 4856 | 0.02 | trans | ||
| rs17294918 | chr16 | 52717122 | NOV | 4856 | 0.09 | trans | ||
| rs16941390 | chr17 | 45050984 | NOV | 4856 | 0.07 | trans | ||
| rs4987765 | chr18 | 60905021 | NOV | 4856 | 0.01 | trans | ||
| rs11873703 | chr18 | 61448702 | NOV | 4856 | 4.187E-4 | trans | ||
| rs17191809 | chr21 | 36444778 | NOV | 4856 | 0.19 | trans | ||
| rs6000401 | chr22 | 37149335 | NOV | 4856 | 0.03 | trans | ||
| rs5991662 | chrX | 43335796 | NOV | 4856 | 0.08 | trans |
Section II. Transcriptome annotation
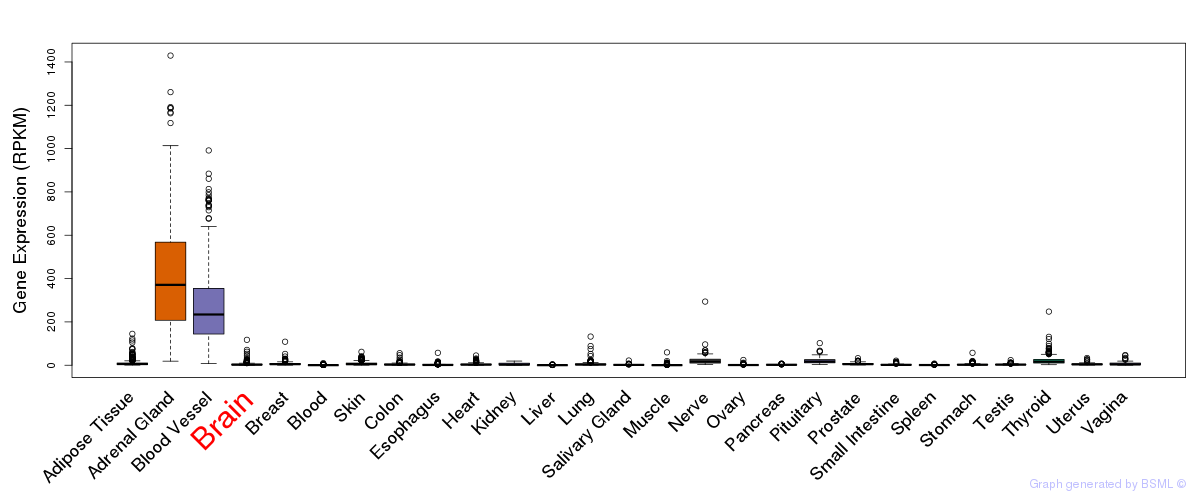
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TAPT1 | 0.60 | 0.58 |
| C1QL1 | 0.60 | 0.66 |
| CPNE8 | 0.59 | 0.69 |
| FGF14 | 0.59 | 0.63 |
| L3MBTL4 | 0.57 | 0.56 |
| SYDE2 | 0.57 | 0.65 |
| CBLN2 | 0.55 | 0.63 |
| NECAB1 | 0.54 | 0.65 |
| NDST3 | 0.54 | 0.57 |
| PGBD5 | 0.53 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IMPA2 | -0.27 | -0.36 |
| RAB13 | -0.27 | -0.36 |
| GNG5 | -0.27 | -0.34 |
| AC098691.2 | -0.27 | -0.29 |
| RFXANK | -0.25 | -0.32 |
| RAB34 | -0.25 | -0.24 |
| C1orf61 | -0.25 | -0.25 |
| H2AFJ | -0.24 | -0.31 |
| GRTP1 | -0.23 | -0.25 |
| IRF7 | -0.23 | -0.32 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WEST ADRENOCORTICAL TUMOR MARKERS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BALDWIN PRKCI TARGETS UP | 35 | 26 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS DN | 35 | 22 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 2WK | 48 | 36 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| STEARMAN TUMOR FIELD EFFECT UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL DN | 18 | 13 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |