Gene Page: NUMA1
Summary ?
| GeneID | 4926 |
| Symbol | NUMA1 |
| Synonyms | NMP-22|NUMA |
| Description | nuclear mitotic apparatus protein 1 |
| Reference | MIM:164009|HGNC:HGNC:8059|Ensembl:ENSG00000137497|HPRD:01236|Vega:OTTHUMG00000167697 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.054 |
| Fetal beta | 0.257 |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0036 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NUMA1 | chr11 | 71725854 | C | T | NM_006185 | p.899A>T | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
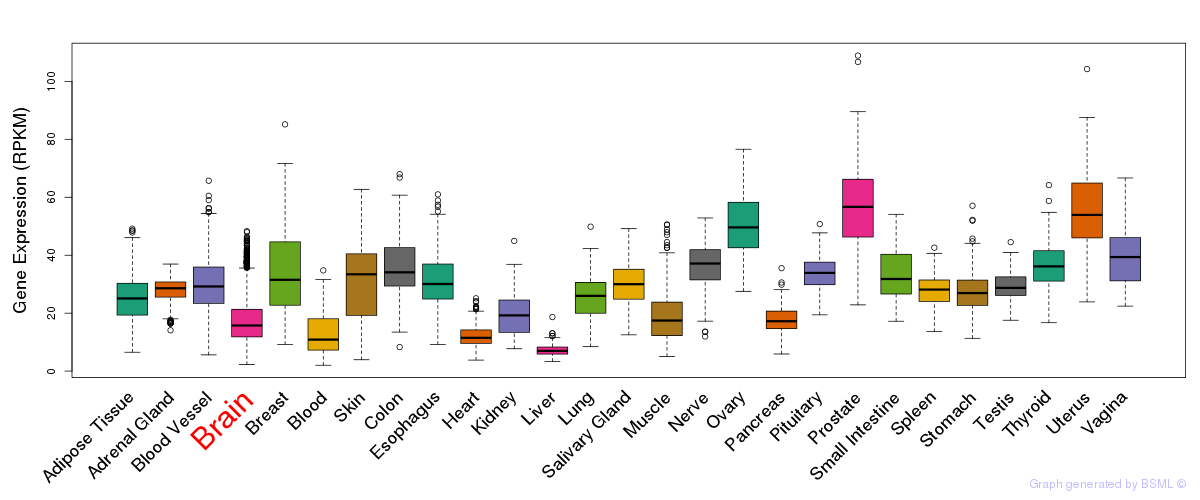
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C4orf8 | 0.92 | 0.95 |
| NASP | 0.91 | 0.91 |
| CLK2 | 0.91 | 0.93 |
| FTSJ3 | 0.91 | 0.93 |
| DNAJC9 | 0.91 | 0.92 |
| RUFY1 | 0.91 | 0.92 |
| PAXIP1 | 0.91 | 0.93 |
| BRD8 | 0.91 | 0.92 |
| XRCC1 | 0.91 | 0.92 |
| SRRT | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.74 | -0.86 |
| AF347015.27 | -0.72 | -0.85 |
| MT-CO2 | -0.71 | -0.84 |
| IFI27 | -0.70 | -0.83 |
| AF347015.33 | -0.69 | -0.81 |
| FXYD1 | -0.69 | -0.81 |
| C5orf53 | -0.68 | -0.72 |
| HLA-F | -0.68 | -0.72 |
| MT-CYB | -0.68 | -0.80 |
| COPZ2 | -0.67 | -0.76 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 15537540 | |
| GO:0005198 | structural molecule activity | TAS | 1541630 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000090 | mitotic anaphase | TAS | 1541636 | |
| GO:0006997 | nucleus organization | TAS | 1541630 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000922 | spindle pole | IDA | 14718566 | |
| GO:0005876 | spindle microtubule | TAS | 1541636 | |
| GO:0005634 | nucleus | TAS | 1541636 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR1A | ARP1 | CTRN1 | FLJ52695 | FLJ52800 | FLJ55002 | ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) | - | HPRD,BioGRID | 10504299 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD,BioGRID | 10189366 |
| EPB41L1 | 4.1N | DKFZp686H17242 | KIAA0338 | MGC11072 | erythrocyte membrane protein band 4.1-like 1 | - | HPRD,BioGRID | 10594058 |
| EPB41L2 | 4.1-G | DKFZp781D1972 | DKFZp781H1755 | erythrocyte membrane protein band 4.1-like 2 | - | HPRD,BioGRID | 12239178 |
| GNAI1 | Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | NuMA interacts with Galphai1. | BIND | 15537540 |
| GPSM2 | LGN | Pins | G-protein signaling modulator 2 (AGS3-like, C. elegans) | - | HPRD,BioGRID | 11781568 |
| GPSM2 | LGN | Pins | G-protein signaling modulator 2 (AGS3-like, C. elegans) | LGN interacts with NuMA. | BIND | 15537540 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | - | HPRD,BioGRID | 12519782 |
| PIM1 | PIM | pim-1 oncogene | - | HPRD,BioGRID | 12111331 |
| RAD21 | FLJ25655 | FLJ40596 | HR21 | HRAD21 | KIAA0078 | MCD1 | NXP1 | SCC1 | hHR21 | RAD21 homolog (S. pombe) | - | HPRD | 11590136 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | - | HPRD,BioGRID | 11590136 |
| SMC3 | BAM | BMH | CDLS3 | CSPG6 | HCAP | SMC3L1 | structural maintenance of chromosomes 3 | Affinity Capture-Western | BioGRID | 11590136 |
| TNKS | PARP-5a | PARP5A | PARPL | TIN1 | TINF1 | TNKS1 | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase | - | HPRD,BioGRID | 12080061 |
| YEATS4 | 4930573H17Rik | B230215M10Rik | GAS41 | NUBI-1 | YAF9 | YEATS domain containing 4 | - | HPRD,BioGRID | 10913114 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RANMS PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREEN DN | 25 | 12 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING DN | 58 | 35 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS DN | 32 | 27 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2 | 39 | 25 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 79 | 85 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-133 | 351 | 357 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-543 | 623 | 629 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.