Gene Page: EVL
Summary ?
| GeneID | 51466 |
| Symbol | EVL |
| Synonyms | RNB6 |
| Description | Enah/Vasp-like |
| Reference | HGNC:HGNC:20234|Ensembl:ENSG00000196405|HPRD:10942|Vega:OTTHUMG00000171530 |
| Gene type | protein-coding |
| Map location | 14q32.2 |
| Pascal p-value | 0.058 |
| Sherlock p-value | 0.1 |
| Fetal beta | 0.089 |
| DMG | 1 (# studies) |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05201677 | 14 | 100606909 | EVL | 1.942E-4 | 0.193 | 0.035 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
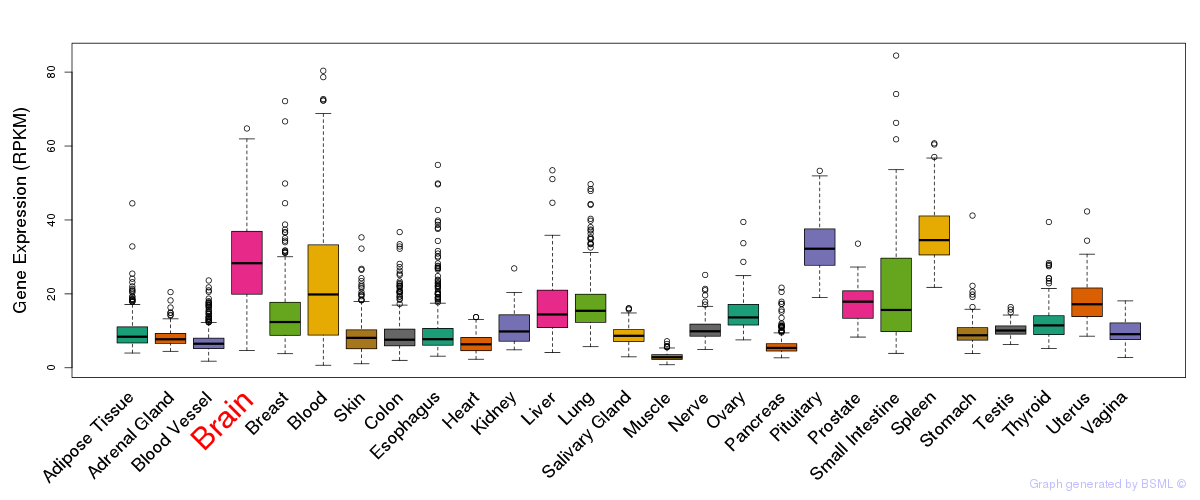
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0005522 | profilin binding | ISS | - | |
| GO:0017124 | SH3 domain binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | NAS | neurite (GO term level: 5) | 10993894 |
| GO:0009887 | organ morphogenesis | NAS | 10993894 | |
| GO:0007166 | cell surface receptor linked signal transduction | NAS | 10993894 | |
| GO:0008154 | actin polymerization or depolymerization | ISS | - | |
| GO:0007015 | actin filament organization | TAS | 9268706 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | ISS | - | |
| GO:0005925 | focal adhesion | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 10945997 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD,BioGRID | 10945997 |
| APBB1IP | INAG1 | PREL1 | RARP1 | RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | RIAM interacts with Evl. | BIND | 15469846 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD | 10945997 |
| DNMBP | KIAA1010 | TUBA | dynamin binding protein | - | HPRD,BioGRID | 14506234 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD | 10945997 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD | 12029088 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 10945997 |
| PFN2 | D3S1319E | PFL | profilin 2 | - | HPRD,BioGRID | 10945997 |
| SEMA6A | HT018 | KIAA1368 | SEMA | SEMA6A1 | SEMAQ | VIA | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | - | HPRD,BioGRID | 10993894 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | AlphaII-spectrin interacts with EVL. | BIND | 15656790 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 10945997 |
| TES | DKFZp586B2022 | MGC1146 | TESS | TESS-2 | testis derived transcript (3 LIM domains) | Tes interacts with EVL. This interaction was modeled on a demonstrated interaction between human Tes and rat EVL. | BIND | 15656790 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERATION OF SECOND MESSENGER MOLECULES | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER UP | 34 | 25 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS DN | 64 | 41 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS UP | 56 | 31 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |