Gene Page: PGD
Summary ?
| GeneID | 5226 |
| Symbol | PGD |
| Synonyms | 6PGD |
| Description | phosphogluconate dehydrogenase |
| Reference | MIM:172200|HGNC:HGNC:8891|Ensembl:ENSG00000142657|HPRD:01391|Vega:OTTHUMG00000001905 |
| Gene type | protein-coding |
| Map location | 1p36.22 |
| Pascal p-value | 0.384 |
| Sherlock p-value | 0.33 |
| Fetal beta | 0.995 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3299 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03344767 | 1 | 10475318 | PGD | 5.113E-4 | 0.275 | 0.047 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16967958 | chr15 | 76483059 | PGD | 5226 | 0.18 | trans |
Section II. Transcriptome annotation
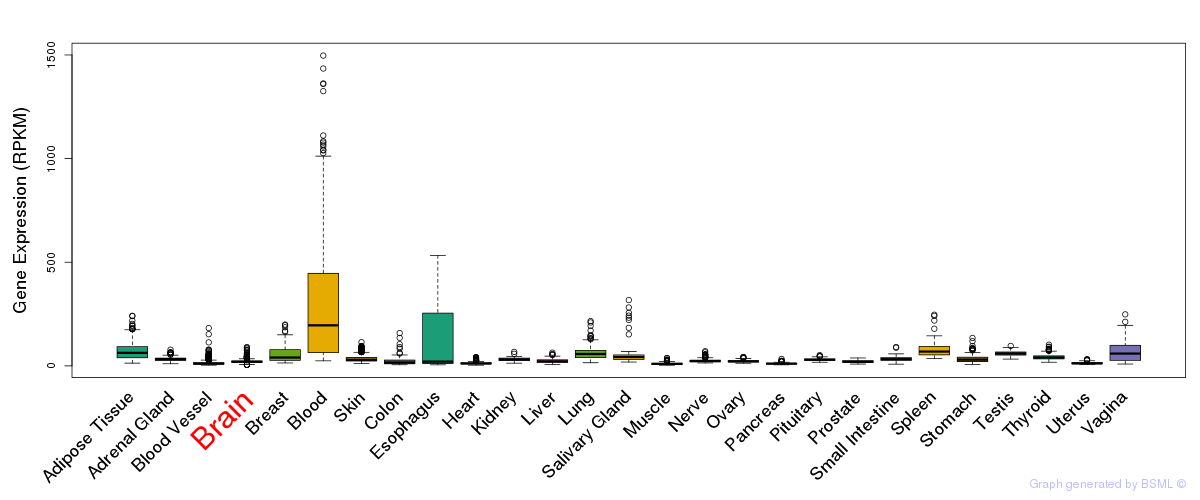
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0050661 | NADP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009051 | pentose-phosphate shunt, oxidative branch | IDA | 3858849 | |
| GO:0019322 | pentose biosynthetic process | IEA | - | |
| GO:0055114 | oxidation reduction | IDA | 3858849 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PENTOSE PHOSPHATE PATHWAY | 27 | 14 | All SZGR 2.0 genes in this pathway |
| KEGG GLUTATHIONE METABOLISM | 50 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA OR RALB TARGETS UP | 48 | 23 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| YAGI AML RELAPSE PROGNOSIS | 35 | 24 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN UP | 16 | 10 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2 | 72 | 52 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID CEBPA NETWORK | 28 | 19 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |