Gene Page: PI4KA
Summary ?
| GeneID | 5297 |
| Symbol | PI4KA |
| Synonyms | PI4K-ALPHA|PIK4CA|PMGYCHA|pi4K230 |
| Description | phosphatidylinositol 4-kinase alpha |
| Reference | MIM:600286|HGNC:HGNC:8983|Ensembl:ENSG00000241973|HPRD:08972|Vega:OTTHUMG00000167440 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.192 |
| Sherlock p-value | 0.376 |
| Fetal beta | -0.93 |
| eGene | Cerebellum |
| Support | CELL METABOLISM G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
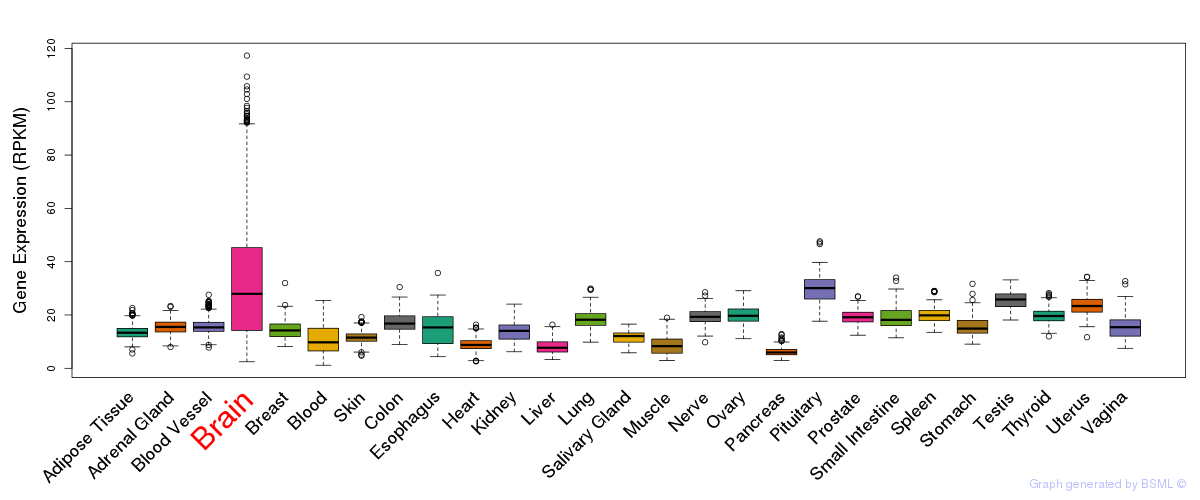
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LTB4R2 | 0.61 | 0.59 |
| LHX2 | 0.59 | 0.56 |
| TMEM145 | 0.59 | 0.64 |
| TMCO6 | 0.58 | 0.64 |
| MOGS | 0.57 | 0.64 |
| TIRAP | 0.57 | 0.69 |
| IL27RA | 0.57 | 0.58 |
| OVCA2 | 0.56 | 0.60 |
| RDH5 | 0.56 | 0.59 |
| MACROD1 | 0.56 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.41 | -0.48 |
| AF347015.27 | -0.40 | -0.47 |
| AF347015.21 | -0.39 | -0.51 |
| AF347015.2 | -0.39 | -0.48 |
| AF347015.8 | -0.38 | -0.45 |
| MT-ATP8 | -0.38 | -0.50 |
| MT-CO2 | -0.38 | -0.44 |
| MT-CYB | -0.38 | -0.45 |
| AF347015.33 | -0.35 | -0.43 |
| AL139819.3 | -0.34 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 14676841 | |
| GO:0004428 | inositol or phosphatidylinositol kinase activity | IEA | - | |
| GO:0004430 | 1-phosphatidylinositol 4-kinase activity | TAS | 8662589 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8662589 |
| GO:0006661 | phosphatidylinositol biosynthetic process | TAS | 7961848 | |
| GO:0007165 | signal transduction | NAS | 7961848 | |
| GO:0048015 | phosphoinositide-mediated signaling | IEA | - | |
| GO:0046854 | phosphoinositide phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005798 | Golgi-associated vesicle | TAS | 8662589 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |