Gene Page: ATP7A
Summary ?
| GeneID | 538 |
| Symbol | ATP7A |
| Synonyms | DSMAX|MK|MNK|SMAX3 |
| Description | ATPase copper transporting alpha |
| Reference | MIM:300011|HGNC:HGNC:869|Ensembl:ENSG00000165240|HPRD:02054|Vega:OTTHUMG00000021885 |
| Gene type | protein-coding |
| Map location | Xq21.1 |
| Fetal beta | 1.155 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 10 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
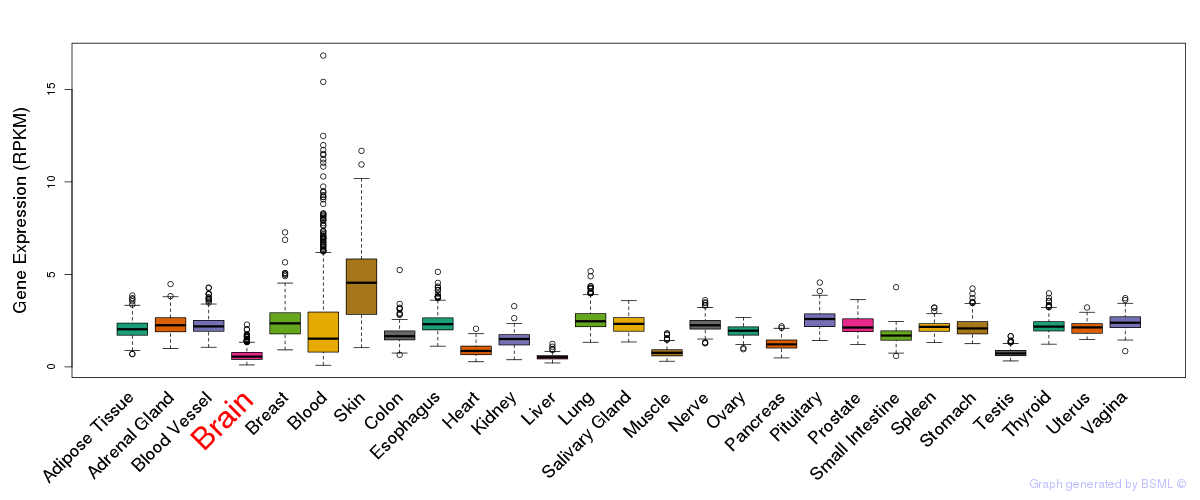
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004008 | copper-exporting ATPase activity | IEA | - | |
| GO:0004008 | copper-exporting ATPase activity | ISS | - | |
| GO:0005375 | copper ion transmembrane transporter activity | IEA | - | |
| GO:0005375 | copper ion transmembrane transporter activity | ISS | - | |
| GO:0005507 | copper ion binding | IDA | 15670166 | |
| GO:0005507 | copper ion binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | TAS | 9817923 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | IEA | - | |
| GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | IEA | - | |
| GO:0016532 | superoxide dismutase copper chaperone activity | ISS | - | |
| GO:0015097 | mercury ion transmembrane transporter activity | IEA | - | |
| GO:0032767 | copper-dependent protein binding | IPI | 10497213 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046873 | metal ion transmembrane transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0021702 | cerebellar Purkinje cell differentiation | ISS | neuron, GABA, Brain (GO term level: 15) | - |
| GO:0021954 | central nervous system neuron development | ISS | neuron (GO term level: 10) | - |
| GO:0021860 | pyramidal neuron development | ISS | neuron, Brain (GO term level: 12) | - |
| GO:0043526 | neuroprotection | ISS | neuron (GO term level: 9) | - |
| GO:0048812 | neurite morphogenesis | ISS | neuron, axon, neurite, dendrite (GO term level: 11) | - |
| GO:0042428 | serotonin metabolic process | ISS | serotonin, Neurotransmitter (GO term level: 8) | - |
| GO:0042417 | dopamine metabolic process | ISS | Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0042415 | norepinephrine metabolic process | ISS | Neurotransmitter (GO term level: 8) | - |
| GO:0001568 | blood vessel development | ISS | - | |
| GO:0002082 | regulation of oxidative phosphorylation | ISS | - | |
| GO:0001974 | blood vessel remodeling | ISS | - | |
| GO:0030001 | metal ion transport | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0010273 | detoxification of copper ion | ISS | - | |
| GO:0007626 | locomotory behavior | ISS | - | |
| GO:0006825 | copper ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0007005 | mitochondrion organization | ISS | - | |
| GO:0006878 | cellular copper ion homeostasis | IMP | 16397091 | |
| GO:0006568 | tryptophan metabolic process | ISS | - | |
| GO:0015694 | mercury ion transport | IEA | - | |
| GO:0015677 | copper ion import | ISS | - | |
| GO:0042093 | T-helper cell differentiation | ISS | - | |
| GO:0018205 | peptidyl-lysine modification | ISS | - | |
| GO:0031069 | hair follicle morphogenesis | ISS | - | |
| GO:0043473 | pigmentation | ISS | - | |
| GO:0030199 | collagen fibril organization | ISS | - | |
| GO:0051216 | cartilage development | ISS | - | |
| GO:0051353 | positive regulation of oxidoreductase activity | IDA | 11092760 | |
| GO:0042414 | epinephrine metabolic process | ISS | - | |
| GO:0048251 | elastic fiber assembly | ISS | - | |
| GO:0048286 | alveolus development | ISS | - | |
| GO:0019430 | removal of superoxide radicals | ISS | - | |
| GO:0043588 | skin development | ISS | - | |
| GO:0048553 | negative regulation of metalloenzyme activity | ISS | - | |
| GO:0048554 | positive regulation of metalloenzyme activity | ISS | - | |
| GO:0051542 | elastin biosynthetic process | ISS | - | |
| GO:0060003 | copper ion export | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | ISS | axon, dendrite (GO term level: 4) | - |
| GO:0043005 | neuron projection | ISS | neuron, axon, neurite, dendrite (GO term level: 5) | - |
| GO:0005794 | Golgi apparatus | IDA | 9467005 | |
| GO:0005802 | trans-Golgi network | IDA | 8943055 |12812980 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005770 | late endosome | IDA | 8943055 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 12812980 | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 8943055 |10567439 |16397091 | |
| GO:0016323 | basolateral plasma membrane | IDA | 16397091 | |
| GO:0030140 | trans-Golgi network transport vesicle | IMP | 9817923 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION TRANSPORT BY P TYPE ATPASES | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KANG CISPLATIN RESISTANCE UP | 19 | 11 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 3363 | 3369 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 522 | 528 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 521 | 528 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-148/152 | 36 | 43 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-182 | 3459 | 3465 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-205 | 2055 | 2061 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-223 | 667 | 674 | 1A,m8 | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-29 | 2060 | 2066 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-5p | 1966 | 1972 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-421 | 3324 | 3330 | 1A | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-9 | 2930 | 2936 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 3459 | 3465 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.