Gene Page: PAQR5
Summary ?
| GeneID | 54852 |
| Symbol | PAQR5 |
| Synonyms | MPRG |
| Description | progestin and adipoQ receptor family member V |
| Reference | MIM:607781|HGNC:HGNC:29645|Ensembl:ENSG00000137819|HPRD:08475|Vega:OTTHUMG00000133322 |
| Gene type | protein-coding |
| Map location | 15q23 |
| Pascal p-value | 0.075 |
| Fetal beta | -0.794 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
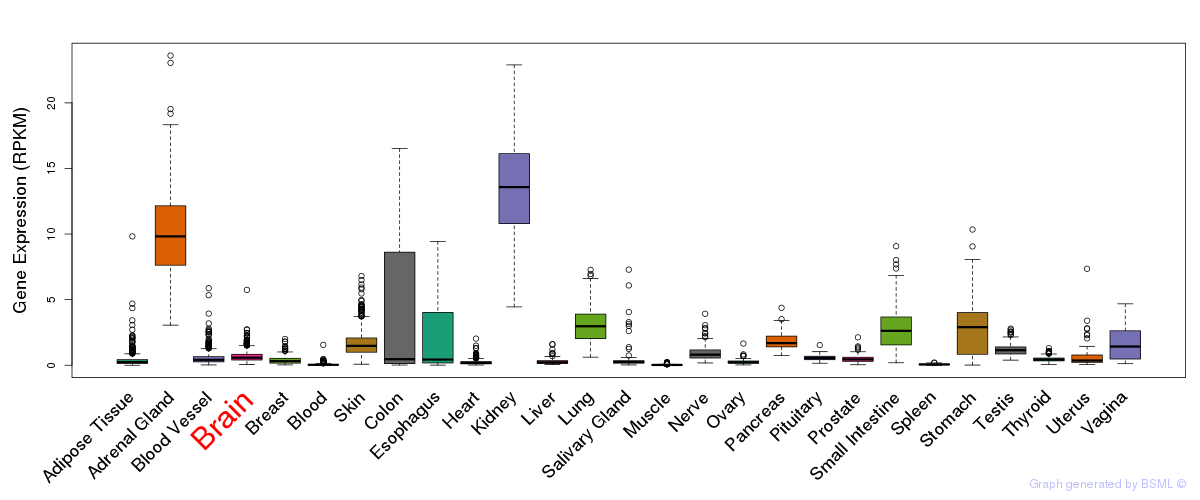
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SFXN3 | 0.88 | 0.88 |
| CAPN5 | 0.87 | 0.87 |
| KIF3C | 0.87 | 0.89 |
| VPS26B | 0.86 | 0.86 |
| NEURL4 | 0.85 | 0.85 |
| ZFYVE27 | 0.85 | 0.86 |
| ST3GAL2 | 0.85 | 0.85 |
| DYNC1I1 | 0.85 | 0.88 |
| IGHMBP2 | 0.84 | 0.84 |
| DTX3 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.76 | -0.76 |
| MT-CO2 | -0.75 | -0.76 |
| AF347015.8 | -0.74 | -0.76 |
| AF347015.2 | -0.74 | -0.75 |
| AF347015.33 | -0.74 | -0.74 |
| MT-CYB | -0.73 | -0.73 |
| AF347015.27 | -0.73 | -0.76 |
| AF347015.15 | -0.72 | -0.73 |
| AF347015.26 | -0.71 | -0.72 |
| AF347015.21 | -0.71 | -0.78 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |