Gene Page: PPP1CB
Summary ?
| GeneID | 5500 |
| Symbol | PPP1CB |
| Synonyms | HEL-S-80p|PP-1B|PP1B|PP1beta|PPP1CD |
| Description | protein phosphatase 1 catalytic subunit beta |
| Reference | MIM:600590|HGNC:HGNC:9282|Ensembl:ENSG00000213639|HPRD:11792|Vega:OTTHUMG00000152011 |
| Gene type | protein-coding |
| Map location | 2p23 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.002 |
| Fetal beta | 0.784 |
| eGene | Myers' cis & trans Meta |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3757318 | chr6 | 151914112 | PPP1CB | 5500 | 0.13 | trans |
Section II. Transcriptome annotation
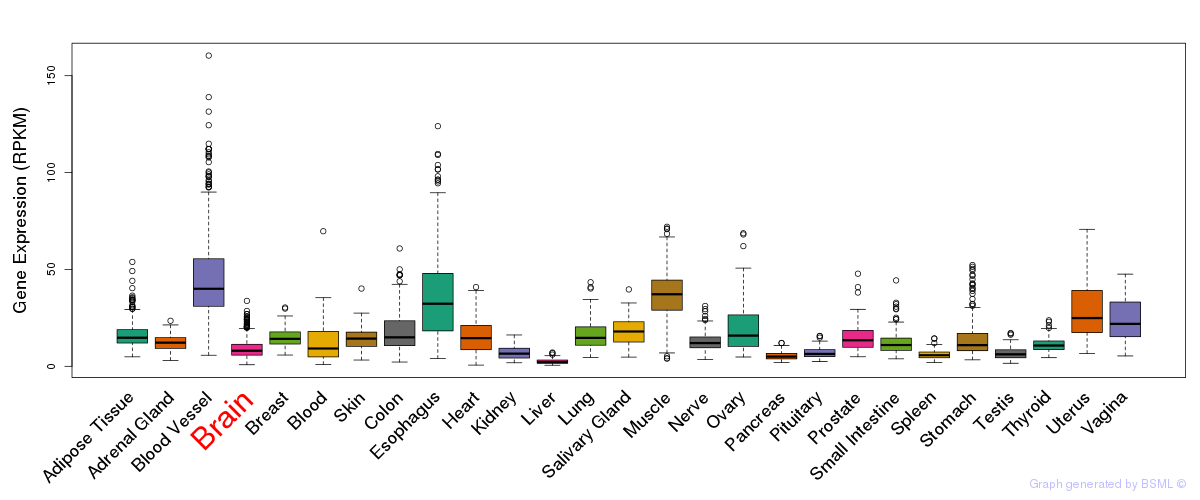
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRUB1 | 0.91 | 0.92 |
| NDFIP1 | 0.90 | 0.90 |
| CALM1 | 0.90 | 0.91 |
| YWHAB | 0.90 | 0.90 |
| RAB6A | 0.90 | 0.90 |
| ITFG1 | 0.90 | 0.90 |
| MTPN | 0.89 | 0.89 |
| ELOVL4 | 0.88 | 0.89 |
| DLAT | 0.87 | 0.87 |
| NECAP1 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.58 | -0.47 |
| EIF4EBP3 | -0.58 | -0.61 |
| GSDMD | -0.58 | -0.57 |
| AP002478.3 | -0.58 | -0.58 |
| AF347015.2 | -0.57 | -0.46 |
| ACSF2 | -0.57 | -0.56 |
| HIGD1B | -0.56 | -0.48 |
| RAB34 | -0.56 | -0.63 |
| FXYD1 | -0.56 | -0.49 |
| MT-CO2 | -0.56 | -0.47 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP11 | AKAP220 | DKFZp781I12161 | FLJ11304 | KIAA0629 | PRKA11 | A kinase (PRKA) anchor protein 11 | Affinity Capture-Western | BioGRID | 12147701 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD | 14743216 |
| NCL | C23 | FLJ45706 | nucleolin | Affinity Capture-Western | BioGRID | 12185196 |
| PPP1R11 | HCG-V | HCGV | MGC125741 | MGC125742 | MGC125743 | TCTE5 | TCTEX5 | protein phosphatase 1, regulatory (inhibitor) subunit 11 | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R12A | MBS | MGC133042 | MYPT1 | protein phosphatase 1, regulatory (inhibitor) subunit 12A | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R12B | MGC131980 | MGC87886 | MYPT2 | protein phosphatase 1, regulatory (inhibitor) subunit 12B | - | HPRD,BioGRID | 9570949 |
| PPP1R12C | DKFZp434D0412 | LENG3 | p84 | protein phosphatase 1, regulatory (inhibitor) subunit 12C | - | HPRD | 11399775 |
| PPP1R15A | GADD34 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | Affinity Capture-Western Phenotypic Suppression | BioGRID | 11564868 |12016208 |
| PPP1R2 | IPP2 | MGC87148 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R7 | SDS22 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R8 | ARD-1 | ARD1 | NIPP-1 | NIPP1 | PRO2047 | protein phosphatase 1, regulatory (inhibitor) subunit 8 | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R9B | FLJ30345 | PPP1R6 | PPP1R9 | SPINO | Spn | protein phosphatase 1, regulatory (inhibitor) subunit 9B | Affinity Capture-Western Reconstituted Complex | BioGRID | 10194355 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 11513739 |
| RYR2 | ARVC2 | ARVD2 | VTSIP | ryanodine receptor 2 (cardiac) | Co-fractionation | BioGRID | 10830164 |
| SH2D4A | FLJ20967 | SH2A | SH2 domain containing 4A | Affinity Capture-MS | BioGRID | 17353931 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-Western Phenotypic Enhancement Reconstituted Complex | BioGRID | 12016208 |
| TMEM33 | 1600019D15Rik | FLJ10525 | SHINC3 | transmembrane protein 33 | Affinity Capture-MS | BioGRID | 17353931 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | - | HPRD | 14640981 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA COAMPLIFIED WITH MYCN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN | 50 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C2 | 18 | 15 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP | 49 | 28 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE UP | 62 | 42 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOGEN METABOLISM | 21 | 16 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |