Gene Page: PRKCE
Summary ?
| GeneID | 5581 |
| Symbol | PRKCE |
| Synonyms | PKCE|nPKC-epsilon |
| Description | protein kinase C epsilon |
| Reference | MIM:176975|HGNC:HGNC:9401|Ensembl:ENSG00000171132|HPRD:01500|Vega:OTTHUMG00000128817 |
| Gene type | protein-coding |
| Map location | 2p21 |
| Pascal p-value | 0.002 |
| Fetal beta | -1.365 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CANABINOID INTRACELLULAR SIGNAL TRANSDUCTION METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 6.2183 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11476211 | 2 | 45879284 | PRKCE | -0.026 | 0.43 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3851333 | chr2 | 45545406 | PRKCE | 5581 | 0.18 | cis | ||
| rs12473388 | chr2 | 45549304 | PRKCE | 5581 | 0.06 | cis | ||
| rs11681887 | chr2 | 45549659 | PRKCE | 5581 | 0.02 | cis | ||
| rs17211786 | chr4 | 182548238 | PRKCE | 5581 | 0.09 | trans | ||
| rs16930362 | chr10 | 74890320 | PRKCE | 5581 | 0.17 | trans |
Section II. Transcriptome annotation
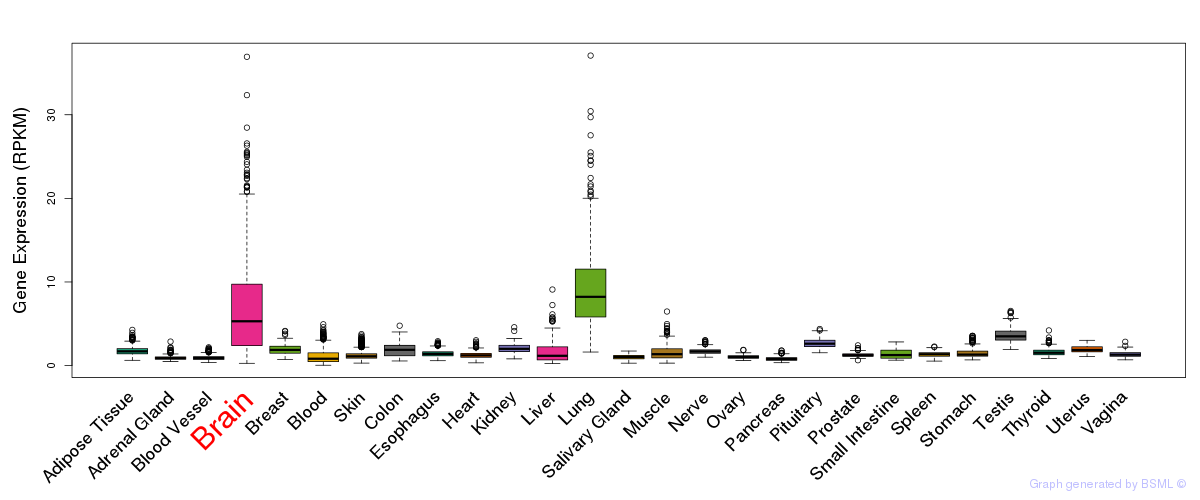
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYT1 | 0.93 | 0.82 |
| PHF21B | 0.91 | 0.84 |
| VANGL2 | 0.91 | 0.89 |
| MED12 | 0.91 | 0.87 |
| CTBP2 | 0.90 | 0.87 |
| SERINC2 | 0.90 | 0.82 |
| NUP188 | 0.90 | 0.89 |
| PLAGL2 | 0.90 | 0.84 |
| FANCC | 0.89 | 0.85 |
| ABL1 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.58 | -0.79 |
| HLA-F | -0.55 | -0.71 |
| FBXO2 | -0.55 | -0.64 |
| PTH1R | -0.54 | -0.66 |
| AIFM3 | -0.54 | -0.69 |
| SLC16A11 | -0.54 | -0.61 |
| LHPP | -0.54 | -0.58 |
| TNFSF12 | -0.53 | -0.61 |
| ALDOC | -0.53 | -0.63 |
| TMEM54 | -0.53 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004871 | signal transducer activity | TAS | 1382605 |10438519 | |
| GO:0004699 | calcium-independent protein kinase C activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 10801826 |17668322 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019992 | diacylglycerol binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 1382605 | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0006917 | induction of apoptosis | TAS | 10438519 | |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 7935319 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11897493 |
| ACTC1 | ACTC | CMD1R | CMH11 | actin, alpha, cardiac muscle 1 | - | HPRD | 11968018 |
| ADAP1 | CENTA1 | GCS1L | p42IP4 | ArfGAP with dual PH domains 1 | Biochemical Activity | BioGRID | 12893243 |
| AFAP1 | AFAP | AFAP-110 | FLJ56849 | actin filament associated protein 1 | - | HPRD,BioGRID | 12134071 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD | 10945988 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | - | HPRD,BioGRID | 11956211 |
| CNN1 | SMCC | Sm-Calp | calponin 1, basic, smooth muscle | - | HPRD | 9312127 |
| COPB1 | COPB | DKFZp761K102 | FLJ10341 | coatomer protein complex, subunit beta 1 | - | HPRD | 11897493 |
| COPB2 | beta'-COP | coatomer protein complex, subunit beta 2 (beta prime) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11897493 |
| COPB2 | beta'-COP | coatomer protein complex, subunit beta 2 (beta prime) | - | HPRD | 9360998 |
| GAD1 | FLJ45882 | GAD | SCP | glutamate decarboxylase 1 (brain, 67kDa) | Biochemical Activity | BioGRID | 15147202 |
| GAD2 | GAD65 | MGC161605 | MGC161607 | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | - | HPRD,BioGRID | 15147202 |
| GJA1 | CX43 | DFNB38 | GJAL | ODDD | gap junction protein, alpha 1, 43kDa | - | HPRD,BioGRID | 10679481 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | Biochemical Activity | BioGRID | 8824244 |
| GNA13 | G13 | MGC46138 | guanine nucleotide binding protein (G protein), alpha 13 | - | HPRD,BioGRID | 8824244 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 11956211 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| HK2 | DKFZp686M1669 | HKII | HXK2 | hexokinase 2 | Affinity Capture-Western | BioGRID | 12663490 |
| KRT1 | CK1 | EHK1 | K1 | KRT1A | keratin 1 | - | HPRD,BioGRID | 11897493 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD,BioGRID | 1374067 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western | BioGRID | 11350735 |
| MYH2 | MYH2A | MYHSA2 | MYHas8 | MyHC-2A | MyHC-IIa | myosin, heavy chain 2, skeletal muscle, adult | - | HPRD | 11897493 |
| MYH9 | DFNA17 | EPSTS | FTNS | MGC104539 | MHA | NMHC-II-A | NMMHCA | myosin, heavy chain 9, non-muscle | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11897493 |
| PDLIM5 | ENH | ENH1 | L9 | LIM | PDZ and LIM domain 5 | - | HPRD,BioGRID | 8940095 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | An unspecified isoform of PDPK1 (PDK1) phosphorylates PRKCE (PKC-epsilon). This interaction was modeled on a demonstrated interaction between PDPK1 from an unspecified species and PRKCE from an unspecified species. | BIND | 12223477 |
| PIK3CB | DKFZp779K1237 | MGC133043 | PI3K | PI3KCB | PI3Kbeta | PIK3C1 | p110-BETA | phosphoinositide-3-kinase, catalytic, beta polypeptide | - | HPRD,BioGRID | 9822674 |
| PPP1R14A | CPI-17 | CPI17 | PPP1INL | protein phosphatase 1, regulatory (inhibitor) subunit 14A | - | HPRD | 15003508 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | PRKCE (PKC-epsilon) autophosphorylates. This interaction was modeled on a demonstrated interaction using PRKCE from an unspecified species. | BIND | 12223477 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | PRKCE (PKC-epsilon) phosphorylates PRKD1 (PKC-mu). This interaction was modeled on a demonstrated interaction between PRKCE from an unspecified species and human PRKD1. | BIND | 12223477 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 11350735 |
| RASGRP3 | GRP3 | KIAA0846 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | Biochemical Activity | BioGRID | 15213298 |
| SLC25A4 | AAC1 | ANT | ANT1 | PEO2 | PEO3 | T1 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | Affinity Capture-Western | BioGRID | 12663490 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 10407019 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 11834516 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 10212259 |
| VDAC1 | MGC111064 | PORIN | PORIN-31-HL | voltage-dependent anion channel 1 | - | HPRD,BioGRID | 12663490 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-Western | BioGRID | 11950841 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID LPA4 PATHWAY | 15 | 10 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR1 PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| PID RAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Z SIGNALLING EVENTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME EFFECTS OF PIP2 HYDROLYSIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL CIS | 65 | 38 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2 | 72 | 52 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 3040 | 3047 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-103/107 | 1564 | 1570 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-141/200a | 1289 | 1296 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-143 | 2180 | 2187 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-144 | 3041 | 3047 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-181 | 3060 | 3066 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 3032 | 3038 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA | ||||
| miR-205 | 2732 | 2739 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-216 | 1495 | 1501 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-218 | 2153 | 2159 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 3057 | 3063 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-25/32/92/363/367 | 1628 | 1635 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-362 | 2735 | 2741 | 1A | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-363 | 2879 | 2885 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-381 | 1503 | 1509 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-500 | 1626 | 1633 | 1A,m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-543 | 1838 | 1844 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-96 | 614 | 620 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.