Gene Page: PRLR
Summary ?
| GeneID | 5618 |
| Symbol | PRLR |
| Synonyms | HPRL|MFAB|hPRLrI |
| Description | prolactin receptor |
| Reference | MIM:176761|HGNC:HGNC:9446|Ensembl:ENSG00000113494|HPRD:01457|Vega:OTTHUMG00000090789 |
| Gene type | protein-coding |
| Map location | 5p13.2 |
| Pascal p-value | 0.203 |
| Fetal beta | -0.582 |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6897259 | 5 | 35188444 | PRLR | ENSG00000113494.12 | 8.942E-7 | 0.01 | 42350 | gtex_brain_putamen_basal |
| rs7721404 | 5 | 35189624 | PRLR | ENSG00000113494.12 | 4.778E-7 | 0.01 | 41170 | gtex_brain_putamen_basal |
| rs9292573 | 5 | 35199433 | PRLR | ENSG00000113494.12 | 6.211E-7 | 0.01 | 31361 | gtex_brain_putamen_basal |
| rs873456 | 5 | 35199657 | PRLR | ENSG00000113494.12 | 3.666E-6 | 0.01 | 31137 | gtex_brain_putamen_basal |
| rs875701 | 5 | 35200656 | PRLR | ENSG00000113494.12 | 9.41E-7 | 0.01 | 30138 | gtex_brain_putamen_basal |
| rs72456163 | 5 | 35201798 | PRLR | ENSG00000113494.12 | 1.523E-6 | 0.01 | 28996 | gtex_brain_putamen_basal |
| rs55902512 | 5 | 35202270 | PRLR | ENSG00000113494.12 | 2.562E-6 | 0.01 | 28524 | gtex_brain_putamen_basal |
| rs2047739 | 5 | 35203338 | PRLR | ENSG00000113494.12 | 1.358E-6 | 0.01 | 27456 | gtex_brain_putamen_basal |
| rs2047740 | 5 | 35203392 | PRLR | ENSG00000113494.12 | 1.23E-6 | 0.01 | 27402 | gtex_brain_putamen_basal |
| rs72734580 | 5 | 35204923 | PRLR | ENSG00000113494.12 | 2.55E-6 | 0.01 | 25871 | gtex_brain_putamen_basal |
| rs75915490 | 5 | 35205592 | PRLR | ENSG00000113494.12 | 2.548E-6 | 0.01 | 25202 | gtex_brain_putamen_basal |
| rs74383180 | 5 | 35209548 | PRLR | ENSG00000113494.12 | 2.913E-6 | 0.01 | 21246 | gtex_brain_putamen_basal |
| rs72734599 | 5 | 35218764 | PRLR | ENSG00000113494.12 | 2.812E-6 | 0.01 | 12030 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
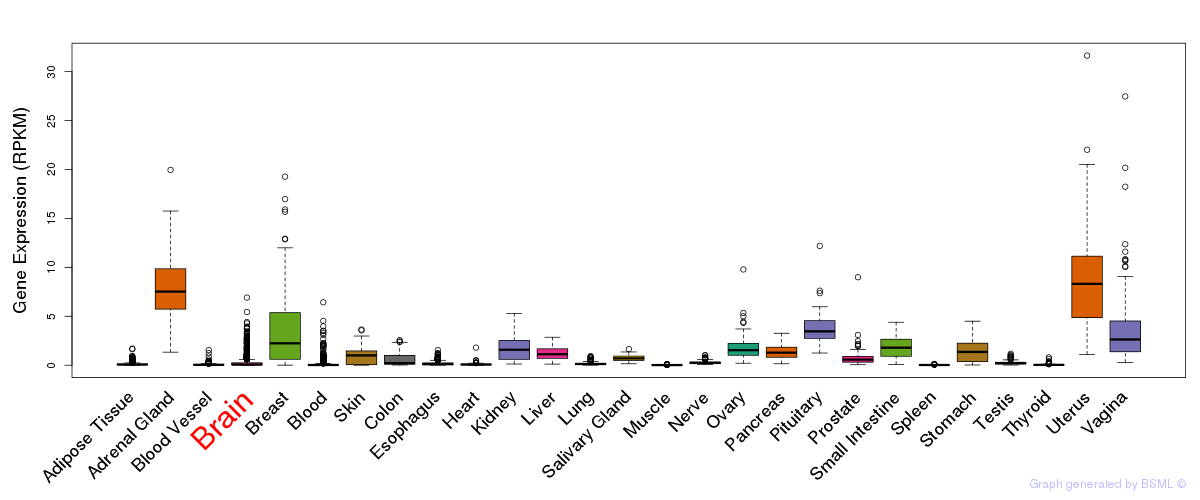
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAPK3 | 0.75 | 0.69 |
| AC019206.3 | 0.71 | 0.66 |
| LRMP | 0.70 | 0.69 |
| PDK2 | 0.69 | 0.68 |
| ANXA11 | 0.67 | 0.84 |
| SLC25A45 | 0.66 | 0.76 |
| ALDOA | 0.66 | 0.76 |
| EXTL1 | 0.65 | 0.82 |
| GJB3 | 0.64 | 0.70 |
| CPEB1 | 0.64 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.39 | -0.57 |
| MMP25 | -0.39 | -0.61 |
| PKN1 | -0.38 | -0.66 |
| KIAA1949 | -0.38 | -0.65 |
| TUBB2B | -0.37 | -0.68 |
| RP9 | -0.37 | -0.64 |
| SH2B2 | -0.37 | -0.69 |
| SH3BP2 | -0.37 | -0.70 |
| YBX1 | -0.37 | -0.66 |
| MARCKSL1 | -0.37 | -0.63 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSH1 | CSA | CSMT | FLJ75407 | PL | chorionic somatomammotropin hormone 1 (placental lactogen) | - | HPRD | 10966654 |
| GH1 | GH | GH-N | GHN | hGH-N | growth hormone 1 | - | HPRD,BioGRID | 9556914 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 10772872 |
| NEK3 | HSPK36 | MGC29949 | NIMA (never in mitosis gene a)-related kinase 3 | Nek3 interacts with PRLr. | BIND | 15618286 |
| PPIA | CYPA | CYPH | MGC117158 | MGC12404 | MGC23397 | peptidylprolyl isomerase A (cyclophilin A) | - | HPRD,BioGRID | 12668872 |
| PRL | - | prolactin | - | HPRD | 9626554 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-Western | BioGRID | 10991949 |
| SOCS2 | CIS2 | Cish2 | SOCS-2 | SSI-2 | SSI2 | STATI2 | suppressor of cytokine signaling 2 | - | HPRD,BioGRID | 9202403 |10455112 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | - | HPRD,BioGRID | 11713228 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | - | HPRD,BioGRID | 11328862 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 7768923 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | Vav2 interacts with PRLr. | BIND | 15618286 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLACTIN RECEPTOR SIGNALING | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES UP | 72 | 38 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| AIYAR COBRA1 TARGETS UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| MATZUK LUTEAL GENES | 8 | 5 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 DN | 40 | 22 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |