Gene Page: SPHK2
Summary ?
| GeneID | 56848 |
| Symbol | SPHK2 |
| Synonyms | SK 2|SK-2|SPK 2|SPK-2 |
| Description | sphingosine kinase 2 |
| Reference | MIM:607092|HGNC:HGNC:18859|Ensembl:ENSG00000063176|HPRD:06157|Vega:OTTHUMG00000183318 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.061 |
| Fetal beta | -0.59 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11376305 | 19 | 49127510 | SPHK2 | 2.56E-5 | -0.44 | 0.017 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
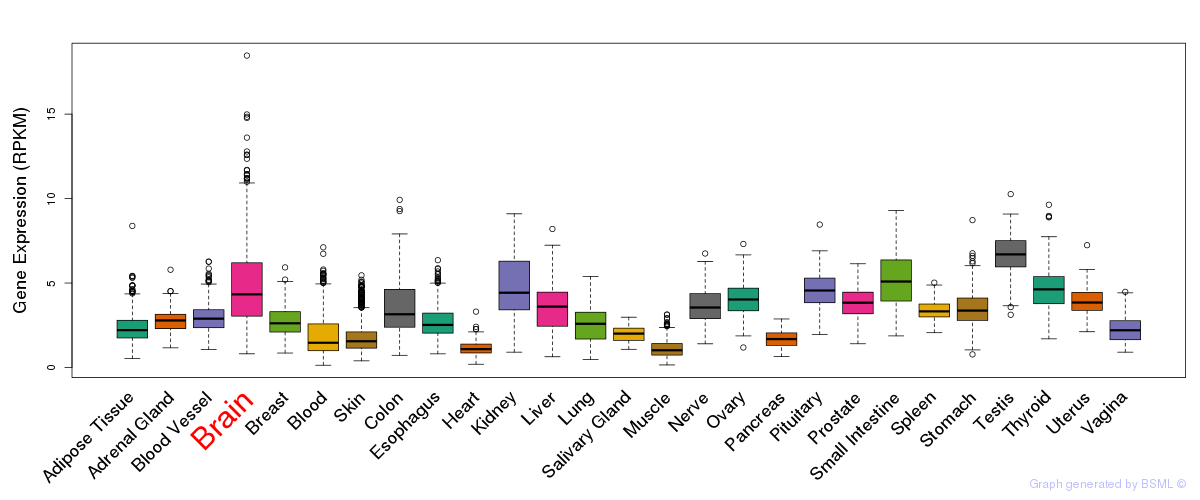
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004143 | diacylglycerol kinase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 11777919 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008481 | sphinganine kinase activity | NAS | 10751414 | |
| GO:0017016 | Ras GTPase binding | NAS | 12391145 | |
| GO:0017050 | D-erythro-sphingosine kinase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0001568 | blood vessel development | IEA | - | |
| GO:0007205 | activation of protein kinase C activity | IEA | - | |
| GO:0008283 | cell proliferation | NAS | 10751414 | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0006669 | sphinganine-1-phosphate biosynthetic process | NAS | 10751414 | |
| GO:0006670 | sphingosine metabolic process | IEA | - | |
| GO:0006916 | anti-apoptosis | NAS | 10751414 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IDA | 10751414 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005624 | membrane fraction | IDA | 10751414 | |
| GO:0005624 | membrane fraction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPHINGOLIPID METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID S1P META PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | 31 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID METABOLISM | 69 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID UP | 25 | 18 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 494 | 501 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-137 | 583 | 589 | m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-153 | 599 | 606 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-19 | 493 | 499 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-25/32/92/363/367 | 602 | 608 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-448 | 600 | 606 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.