Gene Page: PTPN1
Summary ?
| GeneID | 5770 |
| Symbol | PTPN1 |
| Synonyms | PTP1B |
| Description | protein tyrosine phosphatase, non-receptor type 1 |
| Reference | MIM:176885|HGNC:HGNC:9642|Ensembl:ENSG00000196396|HPRD:01477|Vega:OTTHUMG00000032729 |
| Gene type | protein-coding |
| Map location | 20q13.1-q13.2 |
| Pascal p-value | 0.307 |
| Sherlock p-value | 0.778 |
| Fetal beta | 0.228 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0757 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
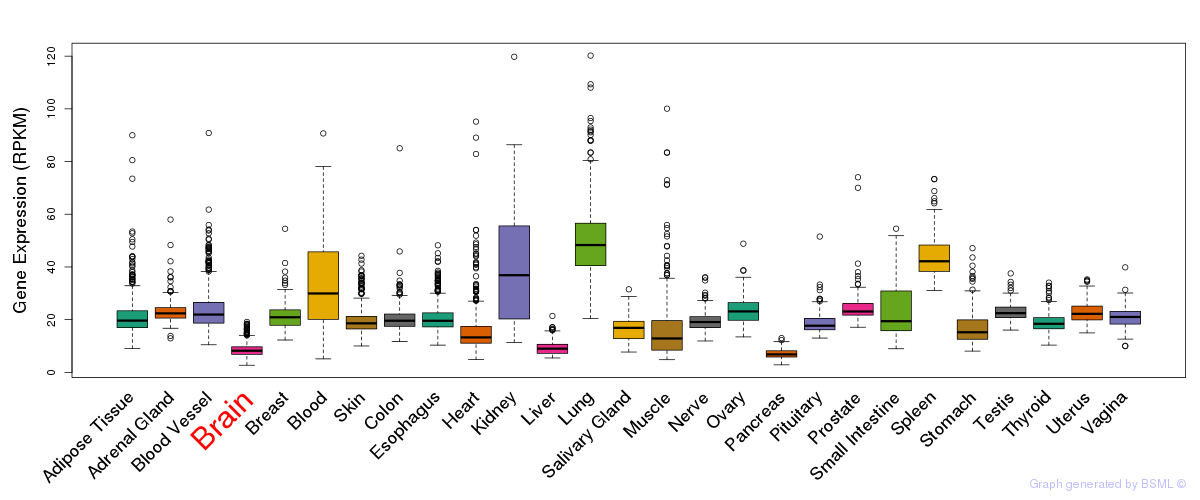
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF142 | 0.94 | 0.95 |
| ZNF384 | 0.94 | 0.96 |
| PFAS | 0.94 | 0.96 |
| HCFC1 | 0.94 | 0.94 |
| DNMT1 | 0.94 | 0.96 |
| ZMIZ1 | 0.94 | 0.95 |
| GCN1L1 | 0.94 | 0.95 |
| KIAA1267 | 0.94 | 0.95 |
| BRPF1 | 0.94 | 0.95 |
| SAP130 | 0.94 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.74 | -0.89 |
| AF347015.27 | -0.72 | -0.88 |
| MT-CO2 | -0.72 | -0.89 |
| AF347015.33 | -0.71 | -0.85 |
| C5orf53 | -0.70 | -0.77 |
| FXYD1 | -0.70 | -0.85 |
| COPZ2 | -0.70 | -0.81 |
| S100B | -0.70 | -0.84 |
| MT-CYB | -0.70 | -0.85 |
| AF347015.8 | -0.69 | -0.87 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11106648 |12907755 |16291744 |17159996 | |
| GO:0004725 | protein tyrosine phosphatase activity | TAS | 10748206 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10748206 | |
| GO:0008286 | insulin receptor signaling pathway | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD | 11579209 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 8940134 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | caveolin-1 interacts with PTP1B. | BIND | 12176037 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 12176037 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | - | HPRD | 12377785 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Reconstituted Complex | BioGRID | 8940134 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 12377785 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 7540771 |8621392 |10889023 |12573287 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 10660596 |12388170 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Reconstituted Complex | BioGRID | 8940134 |10660596 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD,BioGRID | 7514173 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 8999839 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 8826975 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 10660596 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 11694501 |
| LTK | TYK1 | leukocyte receptor tyrosine kinase | - | HPRD,BioGRID | 12237455 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 12237455 |
| NTRK2 | GP145-TrkB | TRKB | neurotrophic tyrosine kinase, receptor, type 2 | - | HPRD,BioGRID | 12237455 |
| NTRK3 | TRKC | gp145(trkC) | neurotrophic tyrosine kinase, receptor, type 3 | - | HPRD,BioGRID | 12237455 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD | 9499405 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD,BioGRID | 11788587 |
| TYK2 | JTK1 | tyrosine kinase 2 | in vivo | BioGRID | 11694501 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNG SIGNALING | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON ALPHA BETA SIGNALING | 64 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNA SIGNALING | 24 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC REGULATORS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS UP | 43 | 24 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 UP | 46 | 29 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 1069 | 1076 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 1764 | 1771 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 1764 | 1770 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-22 | 1426 | 1432 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-299-5p | 1819 | 1825 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.