Gene Page: QDPR
Summary ?
| GeneID | 5860 |
| Symbol | QDPR |
| Synonyms | DHPR|PKU2|SDR33C1 |
| Description | quinoid dihydropteridine reductase |
| Reference | MIM:612676|HGNC:HGNC:9752|Ensembl:ENSG00000151552|HPRD:02025|Vega:OTTHUMG00000128537 |
| Gene type | protein-coding |
| Map location | 4p15.31 |
| Pascal p-value | 0.198 |
| Sherlock p-value | 0.132 |
| Fetal beta | -1.41 |
| eGene | Caudate basal ganglia Cerebellum Putamen basal ganglia |
| Support | RNA AND PROTEIN SYNTHESIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3066730 | 4 | 17483807 | QDPR | ENSG00000151552.7 | 7.705E-7 | 0 | 30050 | gtex_brain_putamen_basal |
| rs2597756 | 4 | 17485081 | QDPR | ENSG00000151552.7 | 1.03E-7 | 0 | 28776 | gtex_brain_putamen_basal |
| rs2597757 | 4 | 17485097 | QDPR | ENSG00000151552.7 | 2.346E-7 | 0 | 28760 | gtex_brain_putamen_basal |
| rs2597758 | 4 | 17485481 | QDPR | ENSG00000151552.7 | 2.38E-7 | 0 | 28376 | gtex_brain_putamen_basal |
| rs2597759 | 4 | 17485644 | QDPR | ENSG00000151552.7 | 3.858E-6 | 0 | 28213 | gtex_brain_putamen_basal |
| rs2597761 | 4 | 17485830 | QDPR | ENSG00000151552.7 | 1.292E-6 | 0 | 28027 | gtex_brain_putamen_basal |
| rs2518626 | 4 | 17485932 | QDPR | ENSG00000151552.7 | 2.428E-7 | 0 | 27925 | gtex_brain_putamen_basal |
| rs2518625 | 4 | 17486029 | QDPR | ENSG00000151552.7 | 2.442E-7 | 0 | 27828 | gtex_brain_putamen_basal |
| rs2518624 | 4 | 17486065 | QDPR | ENSG00000151552.7 | 2.445E-7 | 0 | 27792 | gtex_brain_putamen_basal |
| rs2315247 | 4 | 17486506 | QDPR | ENSG00000151552.7 | 1.995E-7 | 0 | 27351 | gtex_brain_putamen_basal |
| rs2518623 | 4 | 17486575 | QDPR | ENSG00000151552.7 | 2.515E-7 | 0 | 27282 | gtex_brain_putamen_basal |
| rs2597765 | 4 | 17487028 | QDPR | ENSG00000151552.7 | 1.378E-7 | 0 | 26829 | gtex_brain_putamen_basal |
| rs2597766 | 4 | 17487183 | QDPR | ENSG00000151552.7 | 1.414E-7 | 0 | 26674 | gtex_brain_putamen_basal |
| rs12512495 | 4 | 17487597 | QDPR | ENSG00000151552.7 | 2.1E-8 | 0 | 26260 | gtex_brain_putamen_basal |
| rs6449303 | 4 | 17487713 | QDPR | ENSG00000151552.7 | 4.348E-7 | 0 | 26144 | gtex_brain_putamen_basal |
| rs1031326 | 4 | 17488064 | QDPR | ENSG00000151552.7 | 5.947E-7 | 0 | 25793 | gtex_brain_putamen_basal |
| rs699459 | 4 | 17488072 | QDPR | ENSG00000151552.7 | 1.131E-7 | 0 | 25785 | gtex_brain_putamen_basal |
| rs1031327 | 4 | 17488244 | QDPR | ENSG00000151552.7 | 1.131E-7 | 0 | 25613 | gtex_brain_putamen_basal |
| rs699460 | 4 | 17488286 | QDPR | ENSG00000151552.7 | 1.131E-7 | 0 | 25571 | gtex_brain_putamen_basal |
| rs1049582 | 4 | 17488533 | QDPR | ENSG00000151552.7 | 1.131E-7 | 0 | 25324 | gtex_brain_putamen_basal |
| rs1049581 | 4 | 17488534 | QDPR | ENSG00000151552.7 | 1.131E-7 | 0 | 25323 | gtex_brain_putamen_basal |
| rs2597768 | 4 | 17489361 | QDPR | ENSG00000151552.7 | 1.175E-7 | 0 | 24496 | gtex_brain_putamen_basal |
| rs2253130 | 4 | 17489438 | QDPR | ENSG00000151552.7 | 1.144E-7 | 0 | 24419 | gtex_brain_putamen_basal |
| rs2597770 | 4 | 17489695 | QDPR | ENSG00000151552.7 | 1.146E-7 | 0 | 24162 | gtex_brain_putamen_basal |
| rs2252995 | 4 | 17490340 | QDPR | ENSG00000151552.7 | 1.888E-7 | 0 | 23517 | gtex_brain_putamen_basal |
| rs2597771 | 4 | 17491370 | QDPR | ENSG00000151552.7 | 1.17E-7 | 0 | 22487 | gtex_brain_putamen_basal |
| rs731437 | 4 | 17492145 | QDPR | ENSG00000151552.7 | 7.363E-7 | 0 | 21712 | gtex_brain_putamen_basal |
| rs6819744 | 4 | 17497452 | QDPR | ENSG00000151552.7 | 3.593E-6 | 0 | 16405 | gtex_brain_putamen_basal |
| rs2518618 | 4 | 17497861 | QDPR | ENSG00000151552.7 | 4.156E-6 | 0 | 15996 | gtex_brain_putamen_basal |
| rs2597778 | 4 | 17508754 | QDPR | ENSG00000151552.7 | 1.404E-6 | 0 | 5103 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
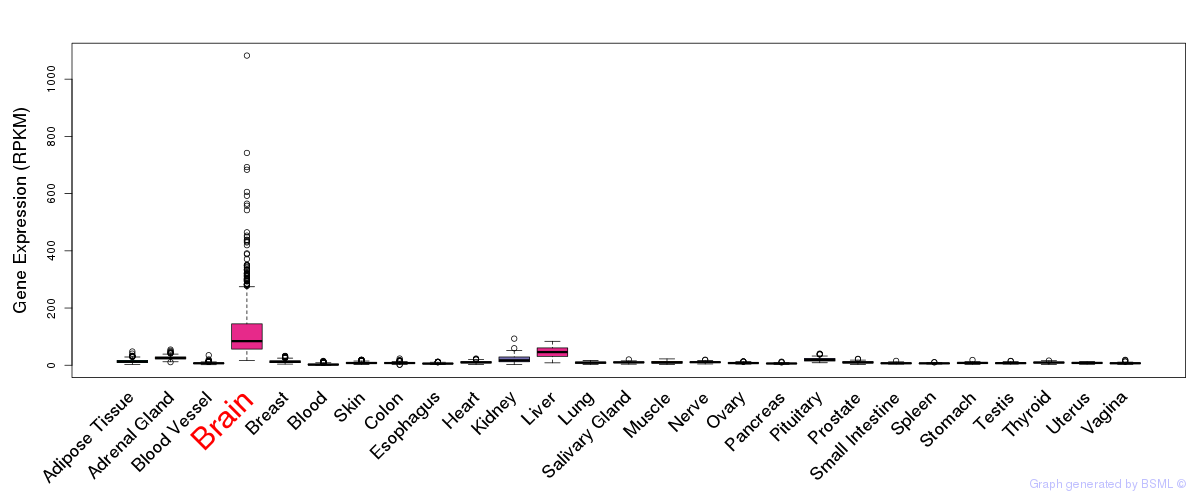
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COBRA1 | 0.89 | 0.87 |
| RIC8A | 0.89 | 0.89 |
| ZMYND19 | 0.88 | 0.88 |
| KLHL22 | 0.88 | 0.88 |
| NLE1 | 0.88 | 0.86 |
| ADPRHL2 | 0.87 | 0.87 |
| BYSL | 0.87 | 0.87 |
| KAT5 | 0.87 | 0.87 |
| DTX3 | 0.87 | 0.87 |
| MED22 | 0.87 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.77 | -0.80 |
| AF347015.8 | -0.76 | -0.81 |
| AF347015.27 | -0.76 | -0.82 |
| AF347015.31 | -0.75 | -0.80 |
| MT-CYB | -0.75 | -0.80 |
| AF347015.33 | -0.74 | -0.78 |
| AF347015.15 | -0.74 | -0.81 |
| AF347015.21 | -0.73 | -0.83 |
| AF347015.2 | -0.72 | -0.77 |
| AF347015.26 | -0.71 | -0.78 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOLATE BIOSYNTHESIS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS UP | 56 | 31 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G12 DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |