Gene Page: RALGDS
Summary ?
| GeneID | 5900 |
| Symbol | RALGDS |
| Synonyms | RGDS|RGF|RalGEF |
| Description | ral guanine nucleotide dissociation stimulator |
| Reference | MIM:601619|HGNC:HGNC:9842|Ensembl:ENSG00000160271|HPRD:03371|Vega:OTTHUMG00000020858 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.195 |
| Fetal beta | 0.799 |
| DMG | 3 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RALGDS | C | T | NM_001042368 | p.T482T | synonymous | NA | NA | Schizophrenia | DNM:Guipponi_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14541011 | 9 | 135989682 | RALGDS | 4.18E-4 | -0.359 | 0.044 | DMG:Wockner_2014 |
| cg17014378 | 9 | 136025100 | RALGDS | 1.44E-9 | -0.021 | 1.39E-6 | DMG:Jaffe_2016 |
| cg18089000 | 9 | 136039706 | RALGDS | 9.898E-4 | 5.14 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2407038 | chr8 | 3588530 | RALGDS | 5900 | 0.14 | trans |
Section II. Transcriptome annotation
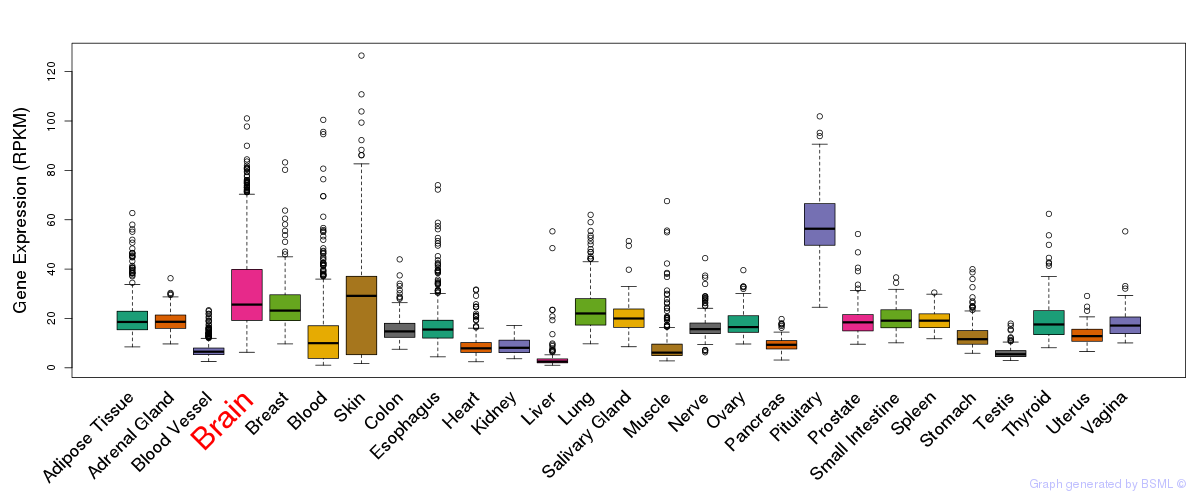
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPC25 | 0.89 | 0.51 |
| KIF11 | 0.89 | 0.54 |
| EXO1 | 0.89 | 0.50 |
| MELK | 0.89 | 0.50 |
| SGOL1 | 0.89 | 0.46 |
| SGOL2 | 0.89 | 0.64 |
| DLGAP5 | 0.88 | 0.46 |
| KIF18A | 0.88 | 0.48 |
| PBK | 0.88 | 0.52 |
| NDC80 | 0.88 | 0.46 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC9A3R2 | -0.36 | -0.40 |
| HLA-F | -0.32 | -0.34 |
| AF347015.27 | -0.31 | -0.35 |
| PTGDS | -0.31 | -0.38 |
| AF347015.15 | -0.31 | -0.36 |
| FBXW4 | -0.31 | -0.37 |
| TINAGL1 | -0.31 | -0.38 |
| MT-CO2 | -0.30 | -0.34 |
| FBXO2 | -0.30 | -0.32 |
| AF347015.33 | -0.30 | -0.36 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARRB1 | ARB1 | ARR1 | arrestin, beta 1 | - | HPRD,BioGRID | 12105416 |
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | - | HPRD,BioGRID | 12105416 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | - | HPRD,BioGRID | 14749388 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Ras interacts with the carboxy terminus of RalGDS. This interaction was modeled on a demonstrated interaction between human Ras and rat RalGDS. | BIND | 9516482 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Reconstituted Complex Two-hybrid | BioGRID | 7809086 |7972015 |9038168 |9150145 |10783161 |10922060 |15031288 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 7972015|9038168 |
| KRAS | C-K-RAS | K-RAS2A | K-RAS2B | K-RAS4A | K-RAS4B | KI-RAS | KRAS1 | KRAS2 | NS3 | RASK2 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | Reconstituted Complex Two-hybrid | BioGRID | 7809086 |10783161 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Two-hybrid | BioGRID | 10783161 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | ERK1 interacts with and phosphorylates RalGDS. | BIND | 9155018 |
| MRAS | FLJ42964 | M-RAs | R-RAS3 | RRAS3 | muscle RAS oncogene homolog | Reconstituted Complex Two-hybrid | BioGRID | 9400994 |10498616 |
| MRAS | FLJ42964 | M-RAs | R-RAS3 | RRAS3 | muscle RAS oncogene homolog | - | HPRD | 9400994|10498616 |
| PDK1 | - | pyruvate dehydrogenase kinase, isozyme 1 | Affinity Capture-Western | BioGRID | 11889038 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD | 11889038 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | - | HPRD,BioGRID | 10085114 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Rap1A interacts with RalGDS. This interaction was modeled on a demonstrated interaction between human Rap1A and RalGDS from an unspecified species. | BIND | 15856025 |
| RAP2A | K-REV | KREV | RAP2 | RbBP-30 | RAP2A, member of RAS oncogene family | - | HPRD,BioGRID | 10085114 |
| RAP2B | MGC20484 | RAP2B, member of RAS oncogene family | - | HPRD,BioGRID | 10085114 |
| RAPGEF2 | CNrasGEF | NRAPGEP | PDZ-GEF1 | PDZGEF1 | RA-GEF | Rap-GEP | Rap guanine nucleotide exchange factor (GEF) 2 | Reconstituted Complex Two-hybrid | BioGRID | 7809086 |
| RIN1 | - | Ras and Rab interactor 1 | Two-hybrid | BioGRID | 10545207 |
| RIT1 | MGC125864 | MGC125865 | RIBB | RIT | ROC1 | Ras-like without CAAX 1 | - | HPRD,BioGRID | 10545207 |
| RIT2 | RIBA | RIN | ROC2 | Ras-like without CAAX 2 | - | HPRD | 10545207 |
| RRAS | - | related RAS viral (r-ras) oncogene homolog | - | HPRD,BioGRID | 7809086 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD,BioGRID | 11788587 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME P38MAPK EVENTS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| SARTIPY BLUNTED BY INSULIN RESISTANCE DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |