Gene Page: BCL2L1
Summary ?
| GeneID | 598 |
| Symbol | BCL2L1 |
| Synonyms | BCL-XL/S|BCL2L|BCLX|Bcl-X|PPP1R52 |
| Description | BCL2 like 1 |
| Reference | MIM:600039|HGNC:HGNC:992|Ensembl:ENSG00000171552|HPRD:02497|Vega:OTTHUMG00000032192 |
| Gene type | protein-coding |
| Map location | 20q11.21 |
| Pascal p-value | 0.81 |
| Sherlock p-value | 0.98 |
| Fetal beta | -0.307 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0455 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
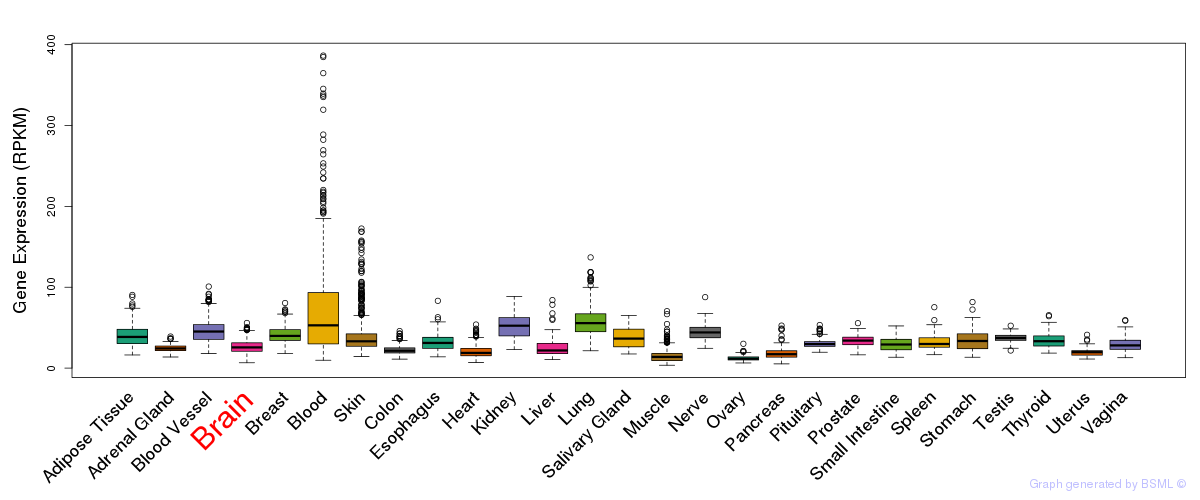
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042802 | identical protein binding | IPI | 15131699 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0001836 | release of cytochrome c from mitochondria | IDA | 9843949 | |
| GO:0008634 | negative regulation of survival gene product expression | TAS | 8358789 | |
| GO:0009314 | response to radiation | IEA | - | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| GO:0045768 | positive regulation of anti-apoptosis | IEA | - | |
| GO:0046902 | regulation of mitochondrial membrane permeability | IDA | 9843949 | |
| GO:0051881 | regulation of mitochondrial membrane potential | IDA | 9843949 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005739 | mitochondrion | IDA | 12011449 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005741 | mitochondrial outer membrane | EXP | 9727492 |10195903 |10198631 |11463392 | |
| GO:0005741 | mitochondrial outer membrane | NAS | 12667443 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0031965 | nuclear membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANTXR1 | ATR | FLJ10601 | FLJ11298 | FLJ21776 | TEM8 | anthrax toxin receptor 1 | - | HPRD | 11574549 |
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | - | HPRD,BioGRID | 9488720 |9539746 |
| AVEN | PDCD12 | apoptosis, caspase activation inhibitor | - | HPRD,BioGRID | 10949025 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD | 7834748 |9305851 |9388232 |11483855 |11494146 |11583631 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Bcl-XL interacts with Bad. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bad. | BIND | 15721256 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Affinity Capture-Western Protein-peptide Reconstituted Complex Two-hybrid | BioGRID | 7834748 |9824152 |10620799 |11206074 |11494146 |12115603 |12137781 |15694340 |15705582 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Bad interacts with Bcl-XL. | BIND | 15694340 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD | 7834748 |11494146|11206074 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Bad interacts with Bcl-xL. | BIND | 15574335 |
| BAG1 | RAP46 | BCL2-associated athanogene | - | HPRD | 10467399 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | - | HPRD,BioGRID | 11175750 |
| BAX | BCL2L4 | BCL2-associated X protein | Bax interacts with Bcl-XL via its BH3 domain. This interaction was modeled on a demonstrated interaction between mouse Bax and human Bcl-XL. | BIND | 11060313 |
| BAX | BCL2L4 | BCL2-associated X protein | Bax interacts with Bcl-XL. | BIND | 9372935 |
| BAX | BCL2L4 | BCL2-associated X protein | Bax interacts with Bcl-xL. | BIND | 15574335 |
| BAX | BCL2L4 | BCL2-associated X protein | Affinity Capture-Western Protein-peptide | BioGRID | 9824152 |12137781 |12853473 |
| BBC3 | JFY1 | PUMA | BCL2 binding component 3 | PUMA interacts with Bcl-xL. | BIND | 15574335 |
| BBC3 | JFY1 | PUMA | BCL2 binding component 3 | Puma interacts with Bcl-XL. | BIND | 15694340 |
| BBC3 | JFY1 | PUMA | BCL2 binding component 3 | Affinity Capture-Western | BioGRID | 15694340 |
| BBC3 | JFY1 | PUMA | BCL2 binding component 3 | Bcl-XL interacts with Puma. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Puma. | BIND | 15721256 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD,BioGRID | 9334338 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 11583631 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Affinity Capture-Western Protein-peptide | BioGRID | 12137781 |14980220 |
| BCL2L10 | BCL-B | Boo | Diva | MGC129810 | MGC129811 | BCL2-like 10 (apoptosis facilitator) | - | HPRD | 11278245 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | - | HPRD | 9430630 |10198631 |11859372|11859372 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Bim interacts with Bcl-XL. | BIND | 15694340 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Affinity Capture-Western FRET | BioGRID | 14596824 |15694340 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | - | HPRD | 11859372 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Bcl-XL interacts with Bim. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bim. | BIND | 15721256 |
| BCL2L14 | BCLG | BCL2-like 14 (apoptosis facilitator) | - | HPRD,BioGRID | 11054413 |
| BCLAF1 | BTF | KIAA0164 | bK211L9.1 | BCL2-associated transcription factor 1 | - | HPRD | 10330179 |
| BECN1 | ATG6 | VPS30 | beclin1 | beclin 1, autophagy related | - | HPRD,BioGRID | 9765397 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Bid interacts with Bcl-XL. | BIND | 15694340 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Protein-peptide | BioGRID | 15694340 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | BCL-XL interacts with the alpha-helical region of the BID-EL BH3 death domain. | BIND | 15353804 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | BCL-XL interacts with the alpha-helical region of the BID BH3 death domain. | BIND | 15353804 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Bcl-XL interacts with Bid. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bid. | BIND | 15721256 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Bid interacts with Bcl-xL. | BIND | 15574335 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | Bik interacts with Bcl-XL. | BIND | 15694340 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | - | HPRD | 9525867 |11342619 |12853473|11342619 |12853473 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | Bcl-XL interacts with Bik. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bik. | BIND | 15721256 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | - | HPRD,BioGRID | 11342619 |12853473 |
| BLK | MGC10442 | B lymphoid tyrosine kinase | Reconstituted Complex | BioGRID | 9525867 |
| BMF | FLJ00065 | Bcl2 modifying factor | Bmf interacts with Bcl-XL. This interaction was modelled on a demonstrated interaction between mouse Bmf and human Bcl-XL. | BIND | 15694340 |
| BMF | FLJ00065 | Bcl2 modifying factor | Bcl-XL interacts with Bmf. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Bmf. | BIND | 15721256 |
| BNIP1 | NIP1 | SEC20 | TRG-8 | BCL2/adenovirus E1B 19kDa interacting protein 1 | - | HPRD,BioGRID | 10217402 |
| BNIP3 | NIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | - | HPRD,BioGRID | 10625696 |
| BNIP3L | BNIP3a | NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like | - | HPRD,BioGRID | 9973195 |10467396|10467396 |
| BNIPL | BNIP-S | BNIPL-1 | BNIPL-2 | BNIPL1 | BNIPL2 | PP753 | BCL2/adenovirus E1B 19kD interacting protein like | Reconstituted Complex Two-hybrid | BioGRID | 9973195 |12901880 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | Phenotypic Suppression | BioGRID | 11832478 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 11733517 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Phenotypic Suppression | BioGRID | 11832478 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9539746 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD,BioGRID | 9326610 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | MRIT-alpha-1 interacts with BclXL. This interaction was modelled on a demonstrated interaction between human MRIT-alpha-1 and BclXL from an unspecified species. | BIND | 9326610 |
| CRYAA | CRYA1 | HSPB4 | crystallin, alpha A | HalphaA-crystallin interacts with Bcl-Xs. | BIND | 14752512 |
| CRYAB | CRYA2 | CTPP2 | HSPB5 | crystallin, alpha B | HalphaB-crystallin interacts with Bcl-Xs. | BIND | 14752512 |
| CYCS | CYC | HCS | cytochrome c, somatic | - | HPRD,BioGRID | 9192670 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-Western | BioGRID | 10198631 |
| FKBP8 | FKBP38 | FKBPr38 | FK506 binding protein 8, 38kDa | - | HPRD,BioGRID | 12510191 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | - | HPRD | 9130713|10075695 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | Hrk interacts with Bcl-XL. | BIND | 15694340 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | Bcl-XL interacts with Hrk. This interaction was modelled on a demonstrated interaction between Bcl-XL from an unspecified species and human Hrk. | BIND | 15721256 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | Hrk interacts with Bcl-XL. | BIND | 9130713 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | - | HPRD,BioGRID | 9130713 |10075695 |
| IKZF3 | AIO | AIOLOS | ZNFN1A3 | IKAROS family zinc finger 3 (Aiolos) | - | HPRD,BioGRID | 11714801 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 10679027 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD | 10679027 |
| MCL1 | BCL2L3 | EAT | MCL1L | MCL1S | MGC104264 | MGC1839 | TM | myeloid cell leukemia sequence 1 (BCL2-related) | Affinity Capture-Western | BioGRID | 10837489 |
| MOAP1 | MAP-1 | PNMA4 | modulator of apoptosis 1 | MAP-1 interacts with Bcl-XL via its amino and carboxy terminal regions. | BIND | 11060313 |
| MOAP1 | MAP-1 | PNMA4 | modulator of apoptosis 1 | - | HPRD,BioGRID | 11060313 |
| PMAIP1 | APR | NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | - | HPRD,BioGRID | 10807576 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | - | HPRD,BioGRID | 12115603 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | - | HPRD,BioGRID | 10446169 |
| PSEN2 | AD3L | AD4 | PS2 | STM2 | presenilin 2 (Alzheimer disease 4) | - | HPRD,BioGRID | 10446169 |
| RAD9A | RAD9 | RAD9 homolog A (S. pombe) | Affinity Capture-Western Two-hybrid | BioGRID | 10620799 |16189514 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Reconstituted Complex | BioGRID | 8929532 |
| RTN1 | MGC133250 | NSP | reticulon 1 | NSP-C interacts with Bcl-XL. | BIND | 11126360 |
| RTN1 | MGC133250 | NSP | reticulon 1 | - | HPRD,BioGRID | 11126360 |
| RTN4 | ASY | NI220/250 | NOGO | NOGO-A | NOGOC | NSP | NSP-CL | Nbla00271 | Nbla10545 | Nogo-B | Nogo-C | RTN-X | RTN4-A | RTN4-B1 | RTN4-B2 | RTN4-C | reticulon 4 | - | HPRD,BioGRID | 11126360 |
| RTN4 | ASY | NI220/250 | NOGO | NOGO-A | NOGOC | NSP | NSP-CL | Nbla00271 | Nbla10545 | Nogo-B | Nogo-C | RTN-X | RTN4-A | RTN4-B1 | RTN4-B2 | RTN4-C | reticulon 4 | RTN-XS interacts with Bcl-XL. | BIND | 11126360 |
| SIVA1 | CD27BP | SIVA | Siva-1 | Siva-2 | SIVA1, apoptosis-inducing factor | - | HPRD | 12011449 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 11742726 |
| SPNS1 | FLJ38358 | HSpin1 | LAT | PP2030 | SPIN1 | SPINL | nrs | spinster homolog 1 (Drosophila) | - | HPRD,BioGRID | 12815463 |
| TMBIM6 | BAXI1 | BI-1 | TEGT | transmembrane BAX inhibitor motif containing 6 | - | HPRD | 9660918 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Bcl-xl interacts with p53. | BIND | 14963330 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 12667443 |
| TPT1 | FLJ27337 | HRF | TCTP | p02 | tumor protein, translationally-controlled 1 | Bcl-xL interacts with TCTP. | BIND | 15870695 |
| VDAC1 | MGC111064 | PORIN | PORIN-31-HL | voltage-dependent anion channel 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10365962 |10737788 |12767928 |15637055 |
| VDAC1 | MGC111064 | PORIN | PORIN-31-HL | voltage-dependent anion channel 1 | - | HPRD | 10049775 |10365962 |10737788 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MITOCHONDRIA PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SA PROGRAMMED CELL DEATH | 12 | 9 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID IL2 STAT5 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLAMMASOMES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| SHANK TAL1 TARGETS DN | 10 | 6 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT UP | 21 | 19 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A | 57 | 36 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| MOOTHA VOXPHOS | 87 | 51 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 1 DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| VARELA ZMPSTE24 TARGETS UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 2 UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D CLUSTER DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 945 | 952 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-133 | 764 | 771 | 1A,m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-142-3p | 88 | 94 | 1A | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-326 | 791 | 797 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-377 | 1441 | 1448 | 1A,m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-491 | 187 | 193 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
| miR-495 | 1233 | 1239 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.