Gene Page: SLC1A6
Summary ?
| GeneID | 6511 |
| Symbol | SLC1A6 |
| Synonyms | EAAT4 |
| Description | solute carrier family 1 member 6 |
| Reference | MIM:600637|HGNC:HGNC:10944|Ensembl:ENSG00000105143|HPRD:09005|Vega:OTTHUMG00000183351 |
| Gene type | protein-coding |
| Map location | 19p13.12 |
| Pascal p-value | 0.445 |
| Sherlock p-value | 0.892 |
| Fetal beta | -0.533 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | METABOTROPIC GLUTAMATE RECEPTOR NEUROTRANSMITTER METABOLISM Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10385290 | 19 | 15083818 | SLC1A6 | 1.662E-4 | 0.703 | 0.033 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6424090 | chr1 | 3628676 | SLC1A6 | 6511 | 0.03 | trans | ||
| rs4950337 | chr1 | 147193821 | SLC1A6 | 6511 | 0.01 | trans | ||
| snp_a-4290143 | 0 | SLC1A6 | 6511 | 0.02 | trans | |||
| rs17572651 | chr1 | 218943612 | SLC1A6 | 6511 | 0.15 | trans | ||
| rs16829545 | chr2 | 151977407 | SLC1A6 | 6511 | 4.003E-9 | trans | ||
| rs16858054 | chr2 | 171105787 | SLC1A6 | 6511 | 0.08 | trans | ||
| rs3845734 | chr2 | 171125572 | SLC1A6 | 6511 | 7.721E-6 | trans | ||
| rs7584986 | chr2 | 184111432 | SLC1A6 | 6511 | 1.137E-11 | trans | ||
| snp_a-1815771 | 0 | SLC1A6 | 6511 | 0.09 | trans | |||
| rs7655318 | chr4 | 153620763 | SLC1A6 | 6511 | 0.16 | trans | ||
| rs2183142 | chr4 | 159232695 | SLC1A6 | 6511 | 0 | trans | ||
| rs17762315 | chr5 | 76807576 | SLC1A6 | 6511 | 0.12 | trans | ||
| rs10491487 | chr5 | 80323367 | SLC1A6 | 6511 | 0.07 | trans | ||
| rs9461864 | chr6 | 33481468 | SLC1A6 | 6511 | 0.01 | trans | ||
| rs1929780 | chr6 | 40757438 | SLC1A6 | 6511 | 0.05 | trans | ||
| rs1929769 | chr6 | 40765977 | SLC1A6 | 6511 | 0.01 | trans | ||
| rs2251379 | chr8 | 73623854 | SLC1A6 | 6511 | 0.05 | trans | ||
| rs16924081 | chr9 | 6169134 | SLC1A6 | 6511 | 0.1 | trans | ||
| rs9406868 | chr9 | 25223372 | SLC1A6 | 6511 | 0.09 | trans | ||
| rs2225105 | chr9 | 25480908 | SLC1A6 | 6511 | 0.03 | trans | ||
| rs11139334 | chr9 | 84209393 | SLC1A6 | 6511 | 0.01 | trans | ||
| rs7036187 | chr9 | 88602015 | SLC1A6 | 6511 | 0.06 | trans | ||
| snp_a-4267673 | 0 | SLC1A6 | 6511 | 0.01 | trans | |||
| rs3765543 | chr9 | 134458323 | SLC1A6 | 6511 | 0.17 | trans | ||
| rs754765 | chr10 | 132013664 | SLC1A6 | 6511 | 0.04 | trans | ||
| rs4076396 | chr11 | 7490235 | SLC1A6 | 6511 | 0.18 | trans | ||
| rs17070045 | chr13 | 58400458 | SLC1A6 | 6511 | 0.18 | trans | ||
| rs17746210 | chr15 | 27882820 | SLC1A6 | 6511 | 0.03 | trans | ||
| rs16955618 | chr15 | 29937543 | SLC1A6 | 6511 | 2.173E-16 | trans | ||
| rs2077735 | chr15 | 58479024 | SLC1A6 | 6511 | 0.09 | trans | ||
| snp_a-4251282 | 0 | SLC1A6 | 6511 | 0.08 | trans | |||
| rs4782029 | chr16 | 17248015 | SLC1A6 | 6511 | 0.17 | trans | ||
| rs16952513 | chr16 | 80165482 | SLC1A6 | 6511 | 0.16 | trans | ||
| rs16945437 | chr17 | 11797650 | SLC1A6 | 6511 | 8.747E-5 | trans | ||
| rs16978352 | chr18 | 42944875 | SLC1A6 | 6511 | 0.03 | trans | ||
| rs17761686 | chr18 | 56647634 | SLC1A6 | 6511 | 0.14 | trans | ||
| rs11882889 | chr19 | 19887684 | SLC1A6 | 6511 | 8.43E-4 | trans | ||
| rs7274477 | chr20 | 1676942 | SLC1A6 | 6511 | 0.11 | trans | ||
| rs1041786 | chr21 | 22617710 | SLC1A6 | 6511 | 0.02 | trans | ||
| rs16986332 | chrX | 10958354 | SLC1A6 | 6511 | 0.07 | trans |
Section II. Transcriptome annotation
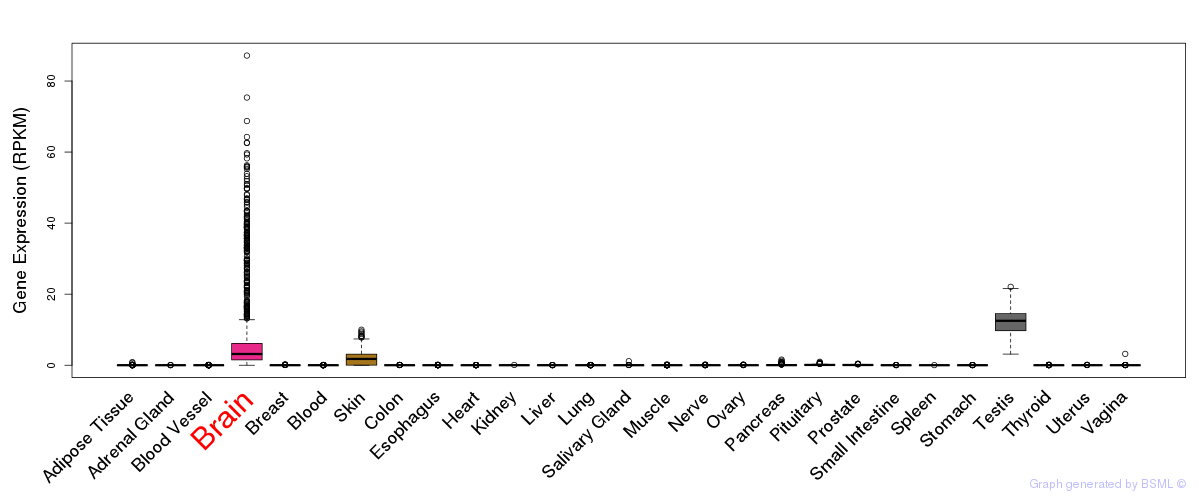
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA0317 | 0.89 | 0.92 |
| TTC9 | 0.87 | 0.90 |
| KIF3C | 0.87 | 0.90 |
| HGSNAT | 0.87 | 0.90 |
| NRXN1 | 0.86 | 0.89 |
| PCDHAC1 | 0.86 | 0.90 |
| MAPT | 0.86 | 0.91 |
| PCYOX1 | 0.86 | 0.88 |
| KLHL18 | 0.85 | 0.91 |
| AC092324.1 | 0.85 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.71 | -0.75 |
| FXYD1 | -0.70 | -0.73 |
| MT-CO2 | -0.69 | -0.74 |
| AC021016.1 | -0.69 | -0.76 |
| AF347015.33 | -0.68 | -0.70 |
| AF347015.27 | -0.67 | -0.72 |
| COPZ2 | -0.67 | -0.71 |
| AF347015.8 | -0.67 | -0.73 |
| HIGD1B | -0.66 | -0.74 |
| MT-CYB | -0.66 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0015183 | L-aspartate transmembrane transporter activity | TAS | glutamate (GO term level: 9) | 7791878 |
| GO:0015293 | symporter activity | IEA | - | |
| GO:0017153 | sodium:dicarboxylate symporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 7791878 |
| GO:0015813 | L-glutamate transport | TAS | glutamate (GO term level: 9) | 7791878 |
| GO:0015810 | aspartate transport | TAS | glutamate (GO term level: 9) | 7791878 |
| GO:0006835 | dicarboxylic acid transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 7791878 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7791878 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | 94 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | 49 | 36 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN | 41 | 29 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |