Gene Page: SMARCE1
Summary ?
| GeneID | 6605 |
| Symbol | SMARCE1 |
| Synonyms | BAF57 |
| Description | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| Reference | MIM:603111|HGNC:HGNC:11109|HPRD:04382| |
| Gene type | protein-coding |
| Map location | 17q21.2 |
| Pascal p-value | 0.749 |
| Sherlock p-value | 0.315 |
| Fetal beta | 1.915 |
| eGene | Myers' cis & trans |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0447 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8074179 | chr17 | 38770964 | SMARCE1 | 6605 | 0.17 | cis | ||
| rs2429548 | chr17 | 38851968 | SMARCE1 | 6605 | 0.05 | cis | ||
| rs2429552 | chr17 | 38868479 | SMARCE1 | 6605 | 0.07 | cis |
Section II. Transcriptome annotation
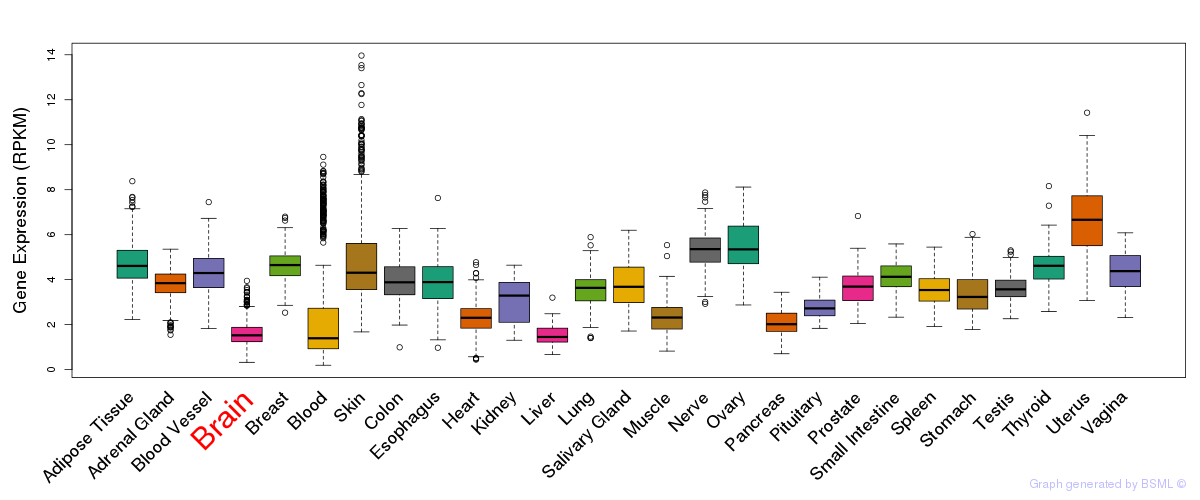
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPP1R12B | 0.85 | 0.84 |
| AP000751.3 | 0.83 | 0.81 |
| HECW2 | 0.82 | 0.82 |
| CDYL2 | 0.82 | 0.80 |
| SIK2 | 0.82 | 0.80 |
| PLEKHM3 | 0.81 | 0.82 |
| ZFP106 | 0.81 | 0.83 |
| CPEB3 | 0.81 | 0.83 |
| DLGAP1 | 0.81 | 0.76 |
| C17orf51 | 0.80 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf61 | -0.48 | -0.57 |
| RHOC | -0.48 | -0.56 |
| AF347015.21 | -0.47 | -0.49 |
| DBI | -0.47 | -0.54 |
| C1orf54 | -0.46 | -0.53 |
| SYCP3 | -0.46 | -0.51 |
| C16orf74 | -0.45 | -0.47 |
| IL32 | -0.44 | -0.53 |
| RAB34 | -0.44 | -0.50 |
| AL050337.1 | -0.44 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003682 | chromatin binding | TAS | 9435219 | |
| GO:0003713 | transcription coactivator activity | TAS | 9435219 | |
| GO:0008080 | N-acetyltransferase activity | IDA | 12192000 | |
| GO:0047485 | protein N-terminus binding | IPI | 12917342 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006337 | nucleosome disassembly | TAS | 9435219 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 9435219 | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 12192000 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000228 | nuclear chromosome | TAS | 9435219 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0017053 | transcriptional repressor complex | IPI | 12192000 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | Affinity Capture-Western | BioGRID | 9845365 |
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | Affinity Capture-Western | BioGRID | 9845365 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD,BioGRID | 12837248 |
| DISC1 | C1orf136 | FLJ13381 | FLJ21640 | FLJ25311 | FLJ41105 | KIAA0457 | SCZD9 | disrupted in schizophrenia 1 | An unspecified isoform of DISC1 interacts with SMARCE1. | BIND | 14623284 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 12145209 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD,BioGRID | 12145209 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Reconstituted Complex | BioGRID | 11018012 |
| KLF1 | EKLF | Kruppel-like factor 1 (erythroid) | Reconstituted Complex | BioGRID | 11018012 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 12145209 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD | 12145209 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 12917342 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 9710619 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | Affinity Capture-MS | BioGRID | 14743216 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Co-purification | BioGRID | 8895581 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Affinity Capture-Western | BioGRID | 9845365 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Affinity Capture-Western | BioGRID | 9845365 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 12145209 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | Affinity Capture-Western | BioGRID | 12244326 |
| TFE3 | RCCP2 | TFEA | bHLHe33 | transcription factor binding to IGHM enhancer 3 | Reconstituted Complex | BioGRID | 11018012 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ERB GENOMIC PATHWAY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 4HR | 54 | 36 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| ASGHARZADEH NEUROBLASTOMA POOR SURVIVAL DN | 46 | 30 | All SZGR 2.0 genes in this pathway |
| GYORFFY MITOXANTRONE RESISTANCE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 292 | 299 | 1A,m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-494 | 1030 | 1036 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-9 | 730 | 736 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.