Gene Page: SON
Summary ?
| GeneID | 6651 |
| Symbol | SON |
| Synonyms | BASS1|C21orf50|DBP-5|NREBP|SON3 |
| Description | SON DNA binding protein |
| Reference | MIM:182465|HGNC:HGNC:11183|Ensembl:ENSG00000159140|HPRD:01678|Vega:OTTHUMG00000065806 |
| Gene type | protein-coding |
| Map location | 21q22.11 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 0.534 |
| Fetal beta | 0.822 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SON | chr21 | 34925493 | G | C | NM_032195 NM_138927 | p.1319G>A p.1319G>A | missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06406294 | 21 | 34915161 | GART;SON | 2.52E-5 | -0.31 | 0.017 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2736751 | chr3 | 60817247 | SON | 6651 | 0.11 | trans |
Section II. Transcriptome annotation
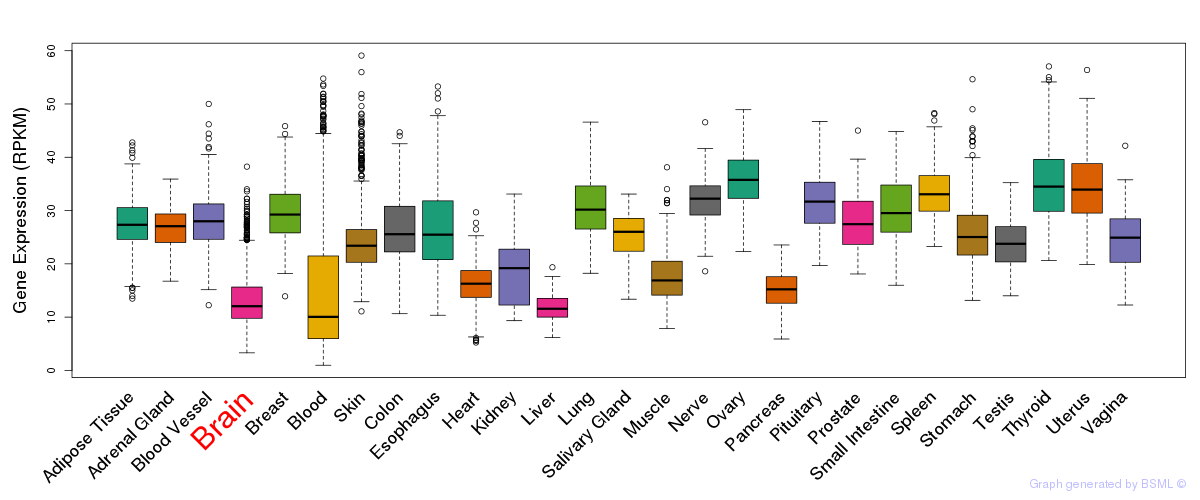
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PVALB | 0.63 | 0.63 |
| DAPL1 | 0.61 | 0.40 |
| SMPX | 0.59 | 0.55 |
| SDR16C5 | 0.59 | 0.47 |
| CPNE9 | 0.58 | 0.57 |
| GNG13 | 0.57 | 0.37 |
| TUBA8 | 0.57 | 0.45 |
| SNCG | 0.56 | 0.60 |
| RDH12 | 0.56 | 0.45 |
| LGALS1 | 0.54 | 0.60 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMURF2 | -0.36 | -0.57 |
| YPEL1 | -0.36 | -0.59 |
| GRAMD4 | -0.36 | -0.48 |
| ZNF589 | -0.35 | -0.56 |
| PPAPR3 | -0.35 | -0.61 |
| TNKS1BP1 | -0.35 | -0.52 |
| CDAN1 | -0.35 | -0.56 |
| STAG3 | -0.35 | -0.50 |
| ZBTB8A | -0.34 | -0.56 |
| VAV2 | -0.34 | -0.58 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |