Gene Page: SPTBN1
Summary ?
| GeneID | 6711 |
| Symbol | SPTBN1 |
| Synonyms | ELF|HEL102|SPTB2|betaSpII |
| Description | spectrin beta, non-erythrocytic 1 |
| Reference | MIM:182790|HGNC:HGNC:11275|Ensembl:ENSG00000115306|HPRD:01683|Vega:OTTHUMG00000133746 |
| Gene type | protein-coding |
| Map location | 2p21 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.407 |
| Fetal beta | 1.019 |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_PocklingtonH1 G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.096 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10186140 | chr2 | 54362974 | SPTBN1 | 6711 | 0.17 | cis | ||
| rs2020214 | chr3 | 73815696 | SPTBN1 | 6711 | 0.07 | trans | ||
| rs11175122 | chr12 | 64084446 | SPTBN1 | 6711 | 0.08 | trans | ||
| rs2717424 | chr12 | 71020775 | SPTBN1 | 6711 | 0.14 | trans | ||
| rs12876724 | chr13 | 67078988 | SPTBN1 | 6711 | 0.1 | trans |
Section II. Transcriptome annotation
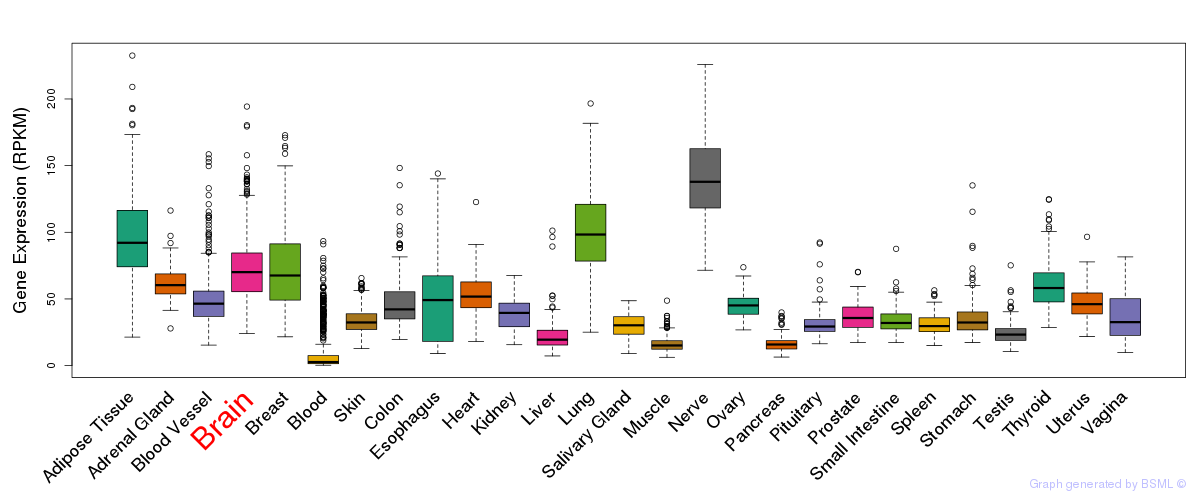
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC4C | 0.78 | 0.87 |
| CDH13 | 0.77 | 0.76 |
| PRPS2 | 0.74 | 0.79 |
| RP4-788L13.1 | 0.72 | 0.67 |
| GPR176 | 0.72 | 0.80 |
| CACNA1I | 0.71 | 0.84 |
| AC012652.1 | 0.71 | 0.82 |
| PCDHA5 | 0.70 | 0.62 |
| MARCH1 | 0.70 | 0.54 |
| FAM19A1 | 0.70 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PECI | -0.39 | -0.38 |
| IMPA2 | -0.37 | -0.45 |
| RAB13 | -0.36 | -0.46 |
| DECR1 | -0.35 | -0.29 |
| ANP32B | -0.34 | -0.46 |
| C11orf67 | -0.33 | -0.26 |
| ACAA2 | -0.32 | -0.29 |
| AK2 | -0.32 | -0.32 |
| AP002478.3 | -0.31 | -0.23 |
| NSBP1 | -0.31 | -0.33 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | TAS | 1527002 | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 1527002 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007184 | SMAD protein nuclear translocation | IEA | - | |
| GO:0007182 | common-partner SMAD protein phosphorylation | IEA | - | |
| GO:0051016 | barbed-end actin filament capping | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 9537418 | |
| GO:0005737 | cytoplasm | IDA | 9537418 | |
| GO:0008091 | spectrin | TAS | 1527002 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR1A | ARP1 | CTRN1 | FLJ52695 | FLJ52800 | FLJ55002 | ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) | - | HPRD | 8991093 |
| CPNE1 | COPN1 | CPN1 | MGC1142 | copine I | - | HPRD | 12522145 |
| CPNE4 | COPN4 | CPN4 | MGC15604 | copine IV | - | HPRD | 12522145 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | Reconstituted Complex | BioGRID | 11069925 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | Affinity Capture-Western | BioGRID | 8991093 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD | 12901833 |
| EPB41L3 | 4.1B | DAL-1 | DAL1 | FLJ37633 | KIAA0987 | erythrocyte membrane protein band 4.1-like 3 | Reconstituted Complex | BioGRID | 15116094 |
| EPB41L3 | 4.1B | DAL-1 | DAL1 | FLJ37633 | KIAA0987 | erythrocyte membrane protein band 4.1-like 3 | EPB41L3 (DAL-1) interacts with an unspecified isoform of SPTBN1 (beta-II-spectrin). | BIND | 15688033 |
| GRIA2 | GLUR2 | GLURB | GluR-K2 | HBGR2 | glutamate receptor, ionotropic, AMPA 2 | Far Western | BioGRID | 10576550 |
| IKBKE | IKK-i | IKKE | IKKI | KIAA0151 | MGC125294 | MGC125295 | MGC125297 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | - | HPRD | 14743216 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 14743216 |
| NF2 | ACN | BANF | SCH | neurofibromin 2 (merlin) | - | HPRD,BioGRID | 11535133 |
| RIPK3 | RIP3 | receptor-interacting serine-threonine kinase 3 | - | HPRD | 14743216 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD | 12543979 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD | 12543979 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | 23 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHOI ATL CHRONIC VS ACUTE DN | 18 | 10 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| ROETH TERT TARGETS DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 10 | 33 | 22 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH EVI1 | 25 | 15 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY DN | 10 | 10 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINBLASTINE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 172 | 178 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-153 | 321 | 327 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 171 | 177 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-219 | 951 | 957 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-410 | 237 | 243 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 321 | 327 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.