Gene Page: BRCA2
Summary ?
| GeneID | 675 |
| Symbol | BRCA2 |
| Synonyms | BRCC2|BROVCA2|FACD|FAD|FAD1|FANCD|FANCD1|GLM3|PNCA2|XRCC11 |
| Description | breast cancer 2 |
| Reference | MIM:600185|HGNC:HGNC:1101|Ensembl:ENSG00000139618|HPRD:02554|Vega:OTTHUMG00000017411 |
| Gene type | protein-coding |
| Map location | 13q12.3 |
| Pascal p-value | 0.808 |
| Fetal beta | 2.008 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0542 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
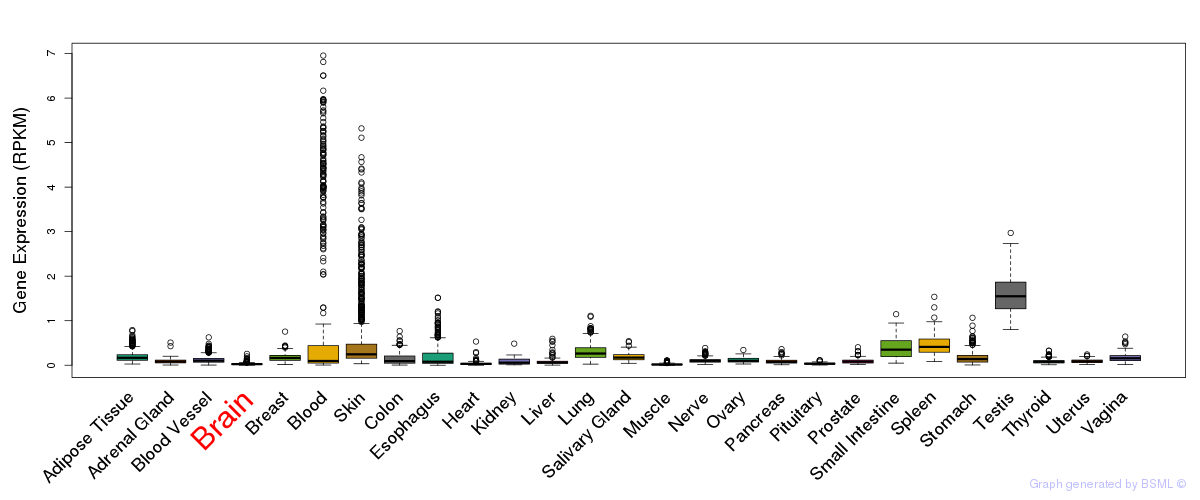
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003697 | single-stranded DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9774970 |10373512 |11597317 |12242698 |15930293 |15967112 |16099937 |16275750 |17515903 | |
| GO:0004402 | histone acetyltransferase activity | IDA | 9824164 | |
| GO:0016563 | transcription activator activity | IDA | 9619837 | |
| GO:0043015 | gamma-tubulin binding | IPI | 17286961 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0001556 | oocyte maturation | IEA | - | |
| GO:0000724 | double-strand break repair via homologous recombination | IEA | - | |
| GO:0001833 | inner cell mass cell proliferation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IC | 9619837 | |
| GO:0007141 | male meiosis I | IEA | - | |
| GO:0006310 | DNA recombination | IEA | - | |
| GO:0008585 | female gonad development | IEA | - | |
| GO:0006289 | nucleotide-excision repair | IMP | 16845393 | |
| GO:0007283 | spermatogenesis | IEA | - | |
| GO:0007090 | regulation of S phase of mitotic cell cycle | NAS | 9774970 | |
| GO:0010225 | response to UV-C | IEA | - | |
| GO:0010165 | response to X-ray | IEA | - | |
| GO:0010332 | response to gamma radiation | IEA | - | |
| GO:0048478 | replication fork protection | IEA | - | |
| GO:0007569 | cell aging | IEA | - | |
| GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | IEA | - | |
| GO:0033595 | response to genistein | IDA | 16434996 | |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | IDA | 15930293 | |
| GO:0033205 | cytokinesis during cell cycle | IDA | 17286961 | |
| GO:0042771 | DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | IEA | - | |
| GO:0043009 | chordate embryonic development | IEA | - | |
| GO:0051298 | centrosome duplication | IDA | 17286961 | |
| GO:0051276 | chromosome organization | IEA | - | |
| GO:0045931 | positive regulation of mitotic cell cycle | IEA | - | |
| GO:0043627 | response to estrogen stimulus | IDA | 8895509 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 17286961 | |
| GO:0005634 | nucleus | IDA | 9560268 |9774970 |17286961 | |
| GO:0005654 | nucleoplasm | EXP | 12606939 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0033593 | BRCA2-MAGE-D1 complex | IDA | 15930293 | |
| GO:0030141 | secretory granule | IDA | 8589722 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Reconstituted Complex | BioGRID | 15199141 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Protein-peptide | BioGRID | 10608806 |
| BARD1 | - | BRCA1 associated RING domain 1 | Co-purification Reconstituted Complex | BioGRID | 14636569 |
| BCCIP | TOK-1 | TOK1 | BRCA2 and CDKN1A interacting protein | - | HPRD,BioGRID | 11313963 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9774970 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with BRCA2. | BIND | 9774970 |
| BRCC3 | BRCC36 | C6.1A | CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BRE | BRCC4 | BRCC45 | brain and reproductive organ-expressed (TNFRSF1A modulator) | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | - | HPRD,BioGRID | 10749118 |
| C11orf30 | EMSY | FLJ90741 | GL002 | chromosome 11 open reading frame 30 | - | HPRD,BioGRID | 14651845 |
| CHEK2 | CDS1 | CHK2 | HuCds1 | LFS2 | PP1425 | RAD53 | CHK2 checkpoint homolog (S. pombe) | Affinity Capture-Western | BioGRID | 17525332 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Affinity Capture-Western Co-localization Two-hybrid | BioGRID | 15115758 |15199141 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | FANCD2 interacts with BRCA2. | BIND | 15115758 |
| FANCE | FACE | FAE | Fanconi anemia, complementation group E | Affinity Capture-Western | BioGRID | 15199141 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | Affinity Capture-Western Two-hybrid | BioGRID | 12915460 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | FANCG interacts with BRCA2. | BIND | 12915460 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 11602572 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | BRCA2 interacts with hsFLNa. | BIND | 11602572 |
| HMG20B | BRAF25 | BRAF35 | FLJ26127 | HMGX2 | PP7706 | SMARCE1r | SOXL | pp8857 | high-mobility group 20B | - | HPRD,BioGRID | 11207365 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD,BioGRID | 9824164 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12815053 |14647413 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Affinity Capture-MS Affinity Capture-Western Co-localization Co-purification Reconstituted Complex Two-hybrid | BioGRID | 9126738 |9405383 |9523196 |9560268 |9774970 |9811893 |11313963 |11477095 |12408834 |12442171 |12815053 |14580352 |14636569 |15065660 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD | 9126738 |12619154 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | BRCA2 interacts with RAD51. | BIND | 9774970 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | BRCA2 interacts with Rad51. | BIND | 9774970 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | BRCA2 interacts with an unspecified isoform of RAD51. | BIND | 15800615 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 12527904 |
| SHFM1 | DSS1 | ECD | SEM1 | SHFD1 | SHSF1 | Shfdg1 | split hand/foot malformation (ectrodactyly) type 1 | - | HPRD,BioGRID | 10373512 |12228710 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Affinity Capture-Western | BioGRID | 12165866 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | SMAD3 interacts with BRCA2. | BIND | 12165866 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 12165866 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 12459499 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9811893 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HOMOLOGOUS RECOMBINATION | 28 | 17 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME FANCONI ANEMIA PATHWAY | 25 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS DN | 139 | 76 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC DN | 45 | 25 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MATHEW FANCONI ANEMIA GENES | 11 | 7 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS UP | 42 | 26 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR UP | 47 | 39 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE LITERATURE | 44 | 25 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |