Gene Page: SYT1
Summary ?
| GeneID | 6857 |
| Symbol | SYT1 |
| Synonyms | P65|SVP65|SYT |
| Description | synaptotagmin 1 |
| Reference | MIM:185605|HGNC:HGNC:11509|Ensembl:ENSG00000067715|HPRD:01710|Vega:OTTHUMG00000134326 |
| Gene type | protein-coding |
| Map location | 12cen-q21 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.923 |
| Fetal beta | -0.706 |
| eGene | Myers' cis & trans |
| Support | CANABINOID DOPAMINE EXOCYTOSIS METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0024 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | SYT1 | 6857 | 0.17 | trans |
Section II. Transcriptome annotation
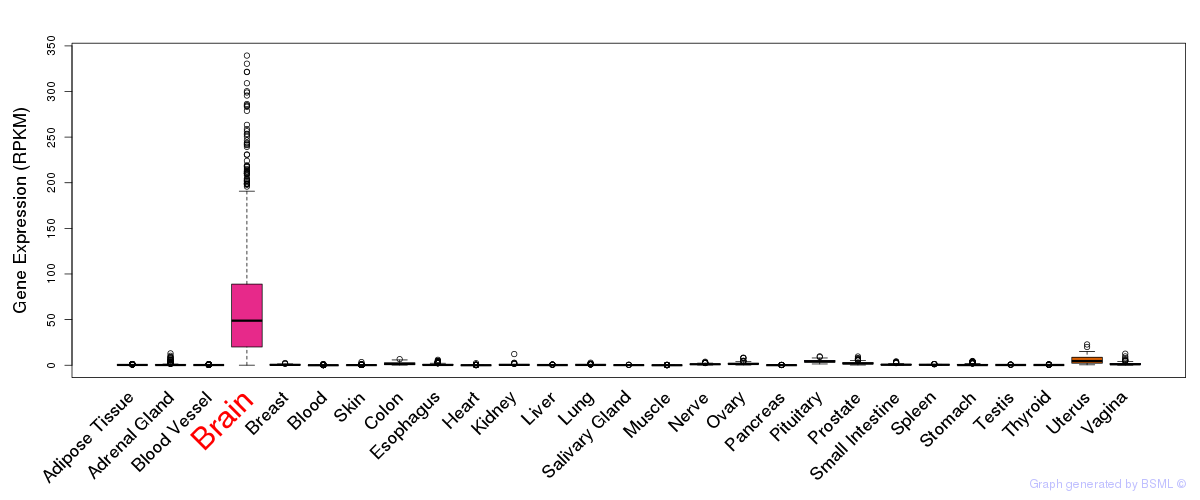
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACTR3 | 0.93 | 0.94 |
| GSPT2 | 0.92 | 0.93 |
| CCT6A | 0.92 | 0.94 |
| SKIV2L2 | 0.92 | 0.93 |
| ARMCX1 | 0.92 | 0.92 |
| SNX4 | 0.92 | 0.92 |
| COPB1 | 0.92 | 0.91 |
| PLAA | 0.92 | 0.93 |
| RAB10 | 0.92 | 0.93 |
| MATR3 | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.73 | -0.85 |
| MT-CO2 | -0.73 | -0.85 |
| AF347015.33 | -0.72 | -0.84 |
| AF347015.31 | -0.71 | -0.83 |
| HLA-F | -0.71 | -0.79 |
| IFI27 | -0.70 | -0.83 |
| AF347015.27 | -0.69 | -0.81 |
| AF347015.8 | -0.69 | -0.83 |
| MT-CYB | -0.68 | -0.81 |
| AF347015.2 | -0.68 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005543 | phospholipid binding | TAS | 1840599 | |
| GO:0005545 | phosphatidylinositol binding | TAS | 11438518 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005215 | transporter activity | IEA | - | |
| GO:0048306 | calcium-dependent protein binding | IEA | - | |
| GO:0017075 | syntaxin-1 binding | TAS | 11438518 | |
| GO:0050750 | low-density lipoprotein receptor binding | IDA | 15082773 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007269 | neurotransmitter secretion | TAS | neuron, axon, Synap, serotonin, Neurotransmitter, dopamine (GO term level: 8) | 11438518 |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 1840599 |
| GO:0005513 | detection of calcium ion | TAS | 11438518 | |
| GO:0006810 | transport | IEA | - | |
| GO:0051260 | protein homooligomerization | TAS | 11438518 | |
| GO:0017157 | regulation of exocytosis | TAS | 11438518 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042734 | presynaptic membrane | IEA | neuron, axon, Synap (GO term level: 5) | - |
| GO:0043005 | neuron projection | IEA | neuron, axon, neurite, dendrite (GO term level: 5) | - |
| GO:0030672 | synaptic vesicle membrane | IEA | Synap (GO term level: 13) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - | |
| GO:0030141 | secretory granule | IEA | - | |
| GO:0030666 | endocytic vesicle membrane | EXP | 2446925 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C6orf15 | STG | chromosome 6 open reading frame 15 | Affinity Capture-Western | BioGRID | 15466855 |
| CACNA1A | APCA | CACNL1A4 | CAV2.1 | EA2 | FHM | HPCA | MHP | MHP1 | SCA6 | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | - | HPRD,BioGRID | 9303303 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | Reconstituted Complex | BioGRID | 8901523 |
| CPLX1 | CPX-I | CPX1 | complexin 1 | - | HPRD | 7553862 |
| FGF1 | AFGF | ECGF | ECGF-beta | ECGFA | ECGFB | FGF-alpha | FGFA | GLIO703 | HBGF1 | fibroblast growth factor 1 (acidic) | - | HPRD,BioGRID | 11432880 |
| GOLM1 | C9orf155 | FLJ22634 | FLJ23608 | GOLPH2 | GP73 | PSEC0257 | bA379P1.3 | golgi membrane protein 1 | Two-hybrid | BioGRID | 16169070 |
| NRXN1 | DKFZp313P2036 | FLJ35941 | Hs.22998 | KIAA0578 | neurexin 1 | - | HPRD | 8901523 |11243866 |
| S100A13 | - | S100 calcium binding protein A13 | - | HPRD,BioGRID | 9712836 |11432880 |12135982 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD,BioGRID | 10692432 |
| STON2 | STN2 | STNB | STNB2 | stonin 2 | - | HPRD,BioGRID | 11381094 |11454741 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD,BioGRID | 9010211 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | Reconstituted Complex | BioGRID | 10397765 |
| STX3 | STX3A | syntaxin 3 | Reconstituted Complex | BioGRID | 10397765 |
| STX4 | STX4A | p35-2 | syntaxin 4 | Reconstituted Complex | BioGRID | 10397765 |
| SV2B | HsT19680 | KIAA0735 | synaptic vesicle glycoprotein 2B | - | HPRD,BioGRID | 15466855 |
| SV2C | MGC118874 | MGC118875 | MGC118876 | MGC118877 | synaptic vesicle glycoprotein 2C | Affinity Capture-Western | BioGRID | 15466855 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | - | HPRD,BioGRID | 10734137 |
| SYT4 | HsT1192 | KIAA1342 | synaptotagmin IV | - | HPRD | 10397765 |
| ZDHHC17 | HIP14 | HIP3 | HSPC294 | HYPH | KIAA0946 | zinc finger, DHHC-type containing 17 | HIP14 catalyzes palmitoylation of Synaptotagmin I. This interaction was modeled on a demonstrated interaction between human HIP14 and Synaptotagmin I from an unspecified species. | BIND | 15603740 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | 17 | 16 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K4ME3 AND H3K27ME3 | 34 | 21 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2 | 98 | 67 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 61 | 67 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 449 | 455 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-128 | 1681 | 1687 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-129-5p | 456 | 462 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-133 | 1602 | 1608 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-135 | 1185 | 1191 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-137 | 216 | 223 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-146 | 1587 | 1594 | 1A,m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-148/152 | 1557 | 1563 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 1624 | 1630 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-19 | 142 | 149 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-193 | 46 | 52 | 1A | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-200bc/429 | 1126 | 1132 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-218 | 1578 | 1584 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-221/222 | 1575 | 1581 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-27 | 1457 | 1463 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-329 | 1572 | 1578 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-34/449 | 1539 | 1545 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-369-3p | 421 | 427 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 421 | 427 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 271 | 277 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-382 | 509 | 516 | 1A,m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-450 | 456 | 462 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-496 | 385 | 391 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.