Gene Page: TAF9
Summary ?
| GeneID | 6880 |
| Symbol | TAF9 |
| Synonyms | MGC:5067|STAF31/32|TAF2G|TAFII-31|TAFII-32|TAFII31|TAFII32|TAFIID32 |
| Description | TATA-box binding protein associated factor 9 |
| Reference | MIM:600822|HGNC:HGNC:11542|Ensembl:ENSG00000085231|Ensembl:ENSG00000273841|HPRD:15983|Vega:OTTHUMG00000099359Vega:OTTHUMG00000187252 |
| Gene type | protein-coding |
| Map location | 5q13.2 |
| Pascal p-value | 0.817 |
| Sherlock p-value | 0.975 |
| Fetal beta | -0.467 |
| eGene | Anterior cingulate cortex BA24 Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TAF9 | chr5 | 68660837 | G | A | NM_001015891 NM_001015892 NM_003187 NM_016283 | . p.243A>V p.243A>V . | intronic missense missense intronic | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1983537 | 0 | TAF9 | 6880 | 0.12 | trans | |||
| rs9885507 | 5 | 68579196 | TAF9 | ENSG00000085231.9 | 4.29318E-6 | 0.04 | 85793 | gtex_brain_ba24 |
| rs71622282 | 5 | 68579200 | TAF9 | ENSG00000085231.9 | 4.30593E-6 | 0.04 | 85789 | gtex_brain_ba24 |
| rs34487969 | 5 | 68579204 | TAF9 | ENSG00000085231.9 | 4.2618E-6 | 0.04 | 85785 | gtex_brain_ba24 |
| rs13356824 | 5 | 68579208 | TAF9 | ENSG00000085231.9 | 4.29314E-6 | 0.04 | 85781 | gtex_brain_ba24 |
Section II. Transcriptome annotation
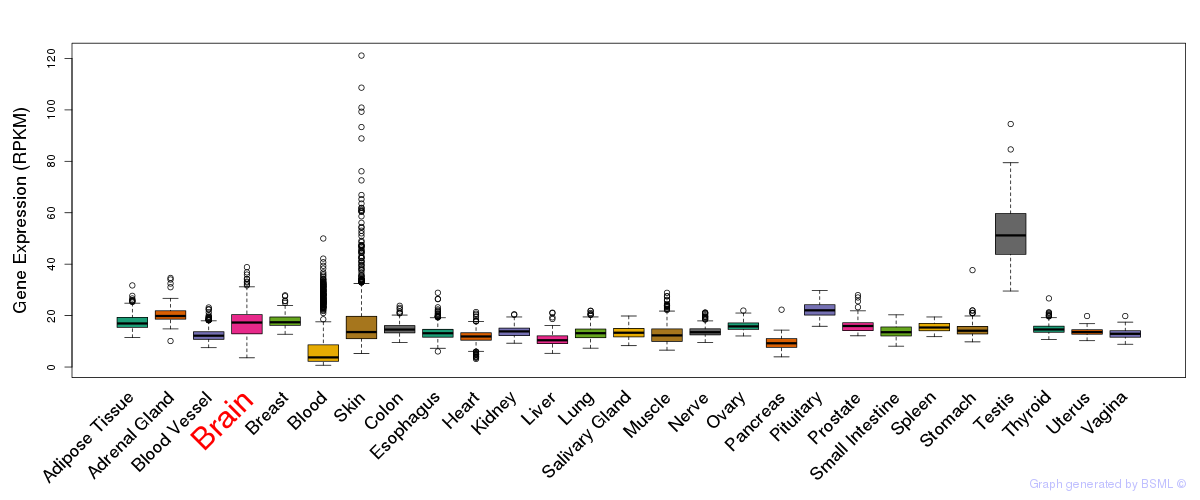
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ECHDC2 | 0.57 | 0.54 |
| MXRA8 | 0.53 | 0.52 |
| LCAT | 0.51 | 0.57 |
| NLRX1 | 0.51 | 0.59 |
| GSX1 | 0.51 | 0.34 |
| EML3 | 0.51 | 0.55 |
| A2ML1 | 0.50 | 0.59 |
| SLC12A4 | 0.50 | 0.60 |
| BCAN | 0.49 | 0.57 |
| C9orf61 | 0.49 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBR1 | -0.43 | -0.33 |
| KLHL1 | -0.42 | -0.30 |
| RLBP1L1 | -0.42 | -0.39 |
| SOX5 | -0.42 | -0.28 |
| AFF3 | -0.42 | -0.33 |
| SEMA3A | -0.41 | -0.32 |
| XPR1 | -0.41 | -0.33 |
| DAB1 | -0.41 | -0.28 |
| BACH2 | -0.41 | -0.27 |
| TIAM2 | -0.41 | -0.33 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C7orf64 | DKFZP564O0523 | DKFZp686D1651 | HSPC304 | chromosome 7 open reading frame 64 | Two-hybrid | BioGRID | 16169070 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD | 10821850 |
| DRAP1 | NC2-alpha | DR1-associated protein 1 (negative cofactor 2 alpha) | Two-hybrid | BioGRID | 16189514 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | - | HPRD | 10821850 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | - | HPRD | 7597030 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | - | HPRD | 7597030 |10821850 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | - | HPRD | 11005381 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | - | HPRD | 10821850 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Affinity Capture-Western | BioGRID | 12660246 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | - | HPRD | 10821850 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 12660246 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD | 10821850 |
| RPS14 | EMTB | ribosomal protein S14 | Two-hybrid | BioGRID | 16189514 |
| SF3B3 | KIAA0017 | RSE1 | SAP130 | SF3b130 | STAF130 | splicing factor 3b, subunit 3, 130kDa | - | HPRD | 11564863 |
| TAF5 | TAF2D | TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa | - | HPRD,BioGRID | 9045704 |
| TAF6 | DKFZp781E21155 | MGC:8964 | TAF2E | TAFII70 | TAFII80 | TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | TAF9 interacts with TAF6. | BIND | 15601843 |
| TAF6 | DKFZp781E21155 | MGC:8964 | TAF2E | TAFII70 | TAFII80 | TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | - | HPRD | 7597030 |7667268 |15601843 |
| TAF6L | FLJ11136 | MGC4288 | PAF65A | TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | - | HPRD | 9674425 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Co-purification | BioGRID | 9153318 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 7761466 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 7761466 |7809597 |11278372 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASAL TRANSCRIPTION FACTORS | 36 | 24 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR DN | 88 | 53 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| NAGY TFTC COMPONENTS HUMAN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| NAGY STAGA COMPONENTS HUMAN | 15 | 8 | All SZGR 2.0 genes in this pathway |
| NAGY PCAF COMPONENTS HUMAN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |