Gene Page: PRDX2
Summary ?
| GeneID | 7001 |
| Symbol | PRDX2 |
| Synonyms | HEL-S-2a|NKEF-B|NKEFB|PRP|PRX2|PRXII|PTX1|TDPX1|TPX1|TSA |
| Description | peroxiredoxin 2 |
| Reference | MIM:600538|HGNC:HGNC:9353|Ensembl:ENSG00000167815|HPRD:02763|Vega:OTTHUMG00000134285 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.045 |
| Sherlock p-value | 0.611 |
| Fetal beta | 0.361 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CELL METABOLISM G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23467004 | 19 | 12912244 | PRDX2 | 1.99E-9 | 0.008 | 1.64E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2105242 | chr1 | 163568328 | PRDX2 | 7001 | 0.19 | trans |
Section II. Transcriptome annotation
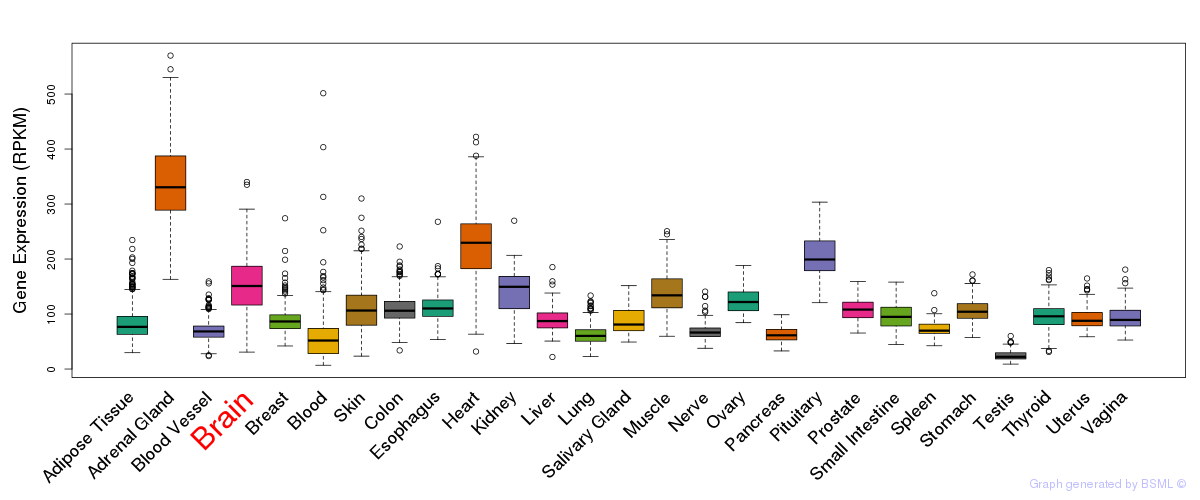
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAP4 | 0.50 | 0.57 |
| TRAF1 | 0.49 | 0.56 |
| TENC1 | 0.47 | 0.58 |
| PPFIBP2 | 0.46 | 0.56 |
| CSDAP1 | 0.44 | 0.41 |
| SERPING1 | 0.44 | 0.54 |
| BCL3 | 0.43 | 0.45 |
| PLAUR | 0.43 | 0.46 |
| ADAMTSL3 | 0.43 | 0.34 |
| C1orf38 | 0.42 | 0.55 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RGS6 | -0.25 | -0.19 |
| SEC62 | -0.25 | -0.25 |
| QRFPR | -0.23 | -0.28 |
| COX6A1 | -0.22 | -0.24 |
| LIN7A | -0.22 | -0.21 |
| CDH7 | -0.22 | -0.12 |
| TCERG1L | -0.22 | -0.12 |
| DOK6 | -0.21 | -0.12 |
| CDH18 | -0.21 | -0.11 |
| IGSF9B | -0.21 | -0.15 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008379 | thioredoxin peroxidase activity | IDA | 8144038 | |
| GO:0016209 | antioxidant activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006979 | response to oxidative stress | IMP | 12943237 | |
| GO:0006979 | response to oxidative stress | NAS | 11904290 | |
| GO:0006916 | anti-apoptosis | TAS | 16130169 | |
| GO:0042981 | regulation of apoptosis | IMP | 12943237 | |
| GO:0045454 | cell redox homeostasis | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 16130169 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| TIMOFEEVA GROWTH STRESS VIA STAT1 DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP | 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIU COMMON CANCER GENES | 79 | 47 | All SZGR 2.0 genes in this pathway |
| RAHMAN TP53 TARGETS PHOSPHORYLATED | 21 | 18 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 MCF10A | 43 | 28 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL CIS | 65 | 38 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST GENETIC VARIANCE | 37 | 21 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| CHIARETTI T ALL REFRACTORY TO THERAPY | 30 | 18 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| BAUS TFF2 TARGETS DN | 11 | 7 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |