Gene Page: THOP1
Summary ?
| GeneID | 7064 |
| Symbol | THOP1 |
| Synonyms | EP24.15|MEPD_HUMAN|MP78|TOP |
| Description | thimet oligopeptidase 1 |
| Reference | MIM:601117|HGNC:HGNC:11793|HPRD:03072| |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.976 |
| Fetal beta | -0.56 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02007638 | 19 | 2785672 | THOP1 | 3.24E-4 | -0.184 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10497672 | 0 | THOP1 | 7064 | 0.2 | trans | |||
| rs11671432 | 19 | 2492758 | THOP1 | ENSG00000172009.10 | 3.24377E-7 | 0.03 | -292700 | gtex_brain_ba24 |
| rs7245952 | 19 | 2498915 | THOP1 | ENSG00000172009.10 | 9.6507E-7 | 0.03 | -286543 | gtex_brain_ba24 |
| rs7245970 | 19 | 2498959 | THOP1 | ENSG00000172009.10 | 9.64999E-7 | 0.03 | -286499 | gtex_brain_ba24 |
Section II. Transcriptome annotation
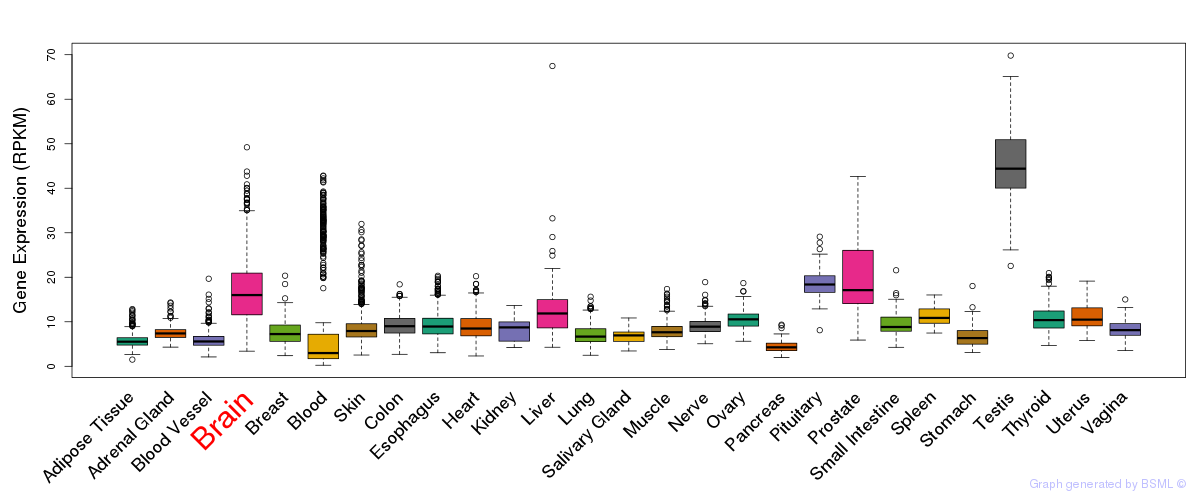
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CYBRD1 | 0.83 | 0.74 |
| GATM | 0.82 | 0.78 |
| SDC2 | 0.82 | 0.71 |
| GLUD1 | 0.81 | 0.74 |
| S1PR1 | 0.81 | 0.71 |
| GJA1 | 0.80 | 0.71 |
| PLSCR4 | 0.80 | 0.77 |
| GLUD2 | 0.79 | 0.73 |
| ADD3 | 0.78 | 0.75 |
| PPP1R3C | 0.78 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF14 | -0.40 | -0.41 |
| MED19 | -0.38 | -0.41 |
| PTMS | -0.38 | -0.42 |
| RTF1 | -0.38 | -0.33 |
| RBMX2 | -0.38 | -0.41 |
| FAM32A | -0.37 | -0.33 |
| RP9P | -0.37 | -0.45 |
| RP9 | -0.37 | -0.39 |
| DPF1 | -0.36 | -0.29 |
| STMN1 | -0.36 | -0.31 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RENIN ANGIOTENSIN SYSTEM | 17 | 10 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE UP | 47 | 32 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA UP | 45 | 30 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |