Gene Page: TNF
Summary ?
| GeneID | 7124 |
| Symbol | TNF |
| Synonyms | DIF|TNF-alpha|TNFA|TNFSF2|TNLG1F |
| Description | tumor necrosis factor |
| Reference | MIM:191160|HGNC:HGNC:11892|Ensembl:ENSG00000232810|HPRD:01855|Vega:OTTHUMG00000031194 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 9.517E-9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
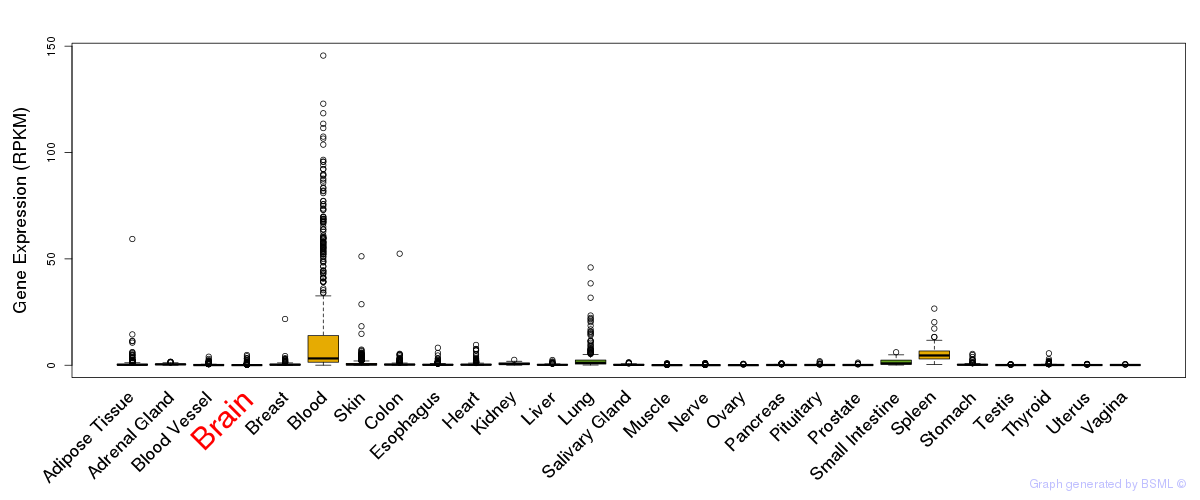
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NUDT18 | 0.90 | 0.85 |
| ACOT8 | 0.89 | 0.92 |
| MECR | 0.89 | 0.91 |
| PGP | 0.89 | 0.90 |
| ALKBH6 | 0.89 | 0.88 |
| TMEM111 | 0.88 | 0.91 |
| DHRS7B | 0.88 | 0.90 |
| KLHDC3 | 0.87 | 0.88 |
| SH3GLB2 | 0.87 | 0.87 |
| FDX1L | 0.87 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| THOC2 | -0.53 | -0.51 |
| EIF5B | -0.52 | -0.62 |
| UPF3A | -0.51 | -0.55 |
| MAP4K4 | -0.51 | -0.47 |
| UPF3B | -0.50 | -0.52 |
| AC005035.1 | -0.50 | -0.47 |
| AC005921.3 | -0.49 | -0.72 |
| ZNF326 | -0.49 | -0.47 |
| RBMX2 | -0.49 | -0.55 |
| KIAA1949 | -0.48 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005164 | tumor necrosis factor receptor binding | IEA | - | |
| GO:0042802 | identical protein binding | IDA | 14512626 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0032800 | receptor biosynthetic process | IDA | Neurotransmitter (GO term level: 5) | 10443688 |
| GO:0000060 | protein import into nucleus, translocation | IDA | 16280327 | |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0000187 | activation of MAPK activity | IDA | 10748004 | |
| GO:0002740 | negative regulation of cytokine secretion during immune response | IDA | 10443688 | |
| GO:0002439 | chronic inflammatory response to antigenic stimulus | IMP | 14512626 | |
| GO:0001934 | positive regulation of protein amino acid phosphorylation | IDA | 10748004 | |
| GO:0008625 | induction of apoptosis via death domain receptors | IEA | - | |
| GO:0006006 | glucose metabolic process | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0006927 | transformed cell apoptosis | IDA | 3883195 | |
| GO:0009615 | response to virus | IDA | 10490959 | |
| GO:0006959 | humoral immune response | IEA | - | |
| GO:0006954 | inflammatory response | IDA | 10748004 | |
| GO:0006919 | caspase activation | IDA | 14512626 | |
| GO:0006916 | anti-apoptosis | IDA | 10748004 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0042346 | positive regulation of NF-kappaB import into nucleus | IDA | 17922812 | |
| GO:0016481 | negative regulation of transcription | IDA | 16895791 | |
| GO:0033209 | tumor necrosis factor-mediated signaling pathway | IMP | 10748004 | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IDA | 16280327 | |
| GO:0051023 | regulation of immunoglobulin secretion | IEA | - | |
| GO:0042127 | regulation of cell proliferation | IEA | - | |
| GO:0050901 | leukocyte tethering or rolling | IDA | 10820279 | |
| GO:0043193 | positive regulation of gene-specific transcription | IDA | 14512626 | |
| GO:0042742 | defense response to bacterium | IEA | - | |
| GO:0032722 | positive regulation of chemokine production | IDA | 10490959 | |
| GO:0032715 | negative regulation of interleukin-6 production | IDA | 10443688 | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEA | - | |
| GO:0045941 | positive regulation of transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0050715 | positive regulation of cytokine secretion | IDA | 10443688 | |
| GO:0045080 | positive regulation of chemokine biosynthetic process | IDA | 10490959 | |
| GO:0045071 | negative regulation of viral genome replication | IDA | 10490959 | |
| GO:0045670 | regulation of osteoclast differentiation | IEA | - | |
| GO:0045994 | positive regulation of translational initiation by iron | IEA | - | |
| GO:0046325 | negative regulation of glucose import | IEA | - | |
| GO:0046330 | positive regulation of JNK cascade | IEA | - | |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | IDA | 16518841 | |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | IDA | 8383325 | |
| GO:0050796 | regulation of insulin secretion | IDA | 8383325 | |
| GO:0051384 | response to glucocorticoid stimulus | IDA | 10443688 | |
| GO:0051798 | positive regulation of hair follicle development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055037 | recycling endosome | ISS | - | |
| GO:0001891 | phagocytic cup | ISS | - | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009897 | external side of plasma membrane | ISS | - | |
| GO:0005886 | plasma membrane | EXP | 2848815 |7758105 |12887920 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BGN | DSPG1 | PG-S1 | PGI | SLRR1A | biglycan | - | HPRD,BioGRID | 12387878 |
| CSF1 | MCSF | MGC31930 | colony stimulating factor 1 (macrophage) | - | HPRD,BioGRID | 8336080 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD,BioGRID | 12387878 |
| KHSRP | FBP2 | FUBP2 | KSRP | MGC99676 | KH-type splicing regulatory protein | KSRP interacts with AREtnf. This interaction was modeled on a demonstrated interaction between KSRP from human and AREtnf from an unspecified species. | BIND | 15175153 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Reconstituted Complex | BioGRID | 12019209 |
| PRTN3 | ACPA | AGP7 | C-ANCA | MBT | P29 | PR-3 | proteinase 3 | - | HPRD,BioGRID | 10339575 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | Affinity Capture-Western | BioGRID | 12887920 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Interaction between TNF (PDB ID: 1A8M_C) and TNF (PDB ID: 1A8M_B). | BIND | 8869635 |9488135 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Interaction between TNF (PDB ID: 1A8M_C) and TNF (PDB ID: 1A8M_A). | BIND | 8869635 |9488135 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Interaction between TNF (PDB ID: 1A8M_B) and TNF (PDB ID: 1A8M_A). | BIND | 8869635 |9488135 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | - | HPRD,BioGRID | 1331108 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD | 1331108 |2158863 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12887920 |14743216 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | TNF-alpha interacts with TNFR1. | BIND | 7852363 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | Affinity Capture-MS | BioGRID | 14743216 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD | 8590321 |9552007 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | TNF-alpha interacts with TNFR-2. | BIND | 7852363 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | Affinity Capture-Western | BioGRID | 12887920 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Affinity Capture-Western | BioGRID | 12887920 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG GRAFT VERSUS HOST DISEASE | 42 | 31 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RELA PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GRANULOCYTES PATHWAY | 14 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDMAC PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LAIR PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TID PATHWAY | 19 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FREE PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL10 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PML PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SODD PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HSP27 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SA MMP CYTOKINE CONNECTION | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PID IL27 PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB CANONICAL PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID ANTHRAX PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB DN | 13 | 10 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 5 | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| RAY ALZHEIMERS DISEASE | 13 | 7 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| BUDHU LIVER CANCER METASTASIS DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K27ME3 | 74 | 46 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K27ME3 | 54 | 32 | All SZGR 2.0 genes in this pathway |
| SEIKE LUNG CANCER POOR SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K27ME3 | 42 | 27 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |