Gene Page: UCHL1
Summary ?
| GeneID | 7345 |
| Symbol | UCHL1 |
| Synonyms | HEL-117|HEL-S-53|NDGOA|PARK5|PGP 9.5|PGP9.5|PGP95|Uch-L1 |
| Description | ubiquitin C-terminal hydrolase L1 |
| Reference | MIM:191342|HGNC:HGNC:12513|Ensembl:ENSG00000154277|HPRD:01877|Vega:OTTHUMG00000099377 |
| Gene type | protein-coding |
| Map location | 4p14 |
| Pascal p-value | 0.927 |
| Sherlock p-value | 0.563 |
| Fetal beta | 0.181 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
| Support | RNA AND PROTEIN SYNTHESIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0667 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | UCHL1 | 7345 | 0.1 | trans |
Section II. Transcriptome annotation
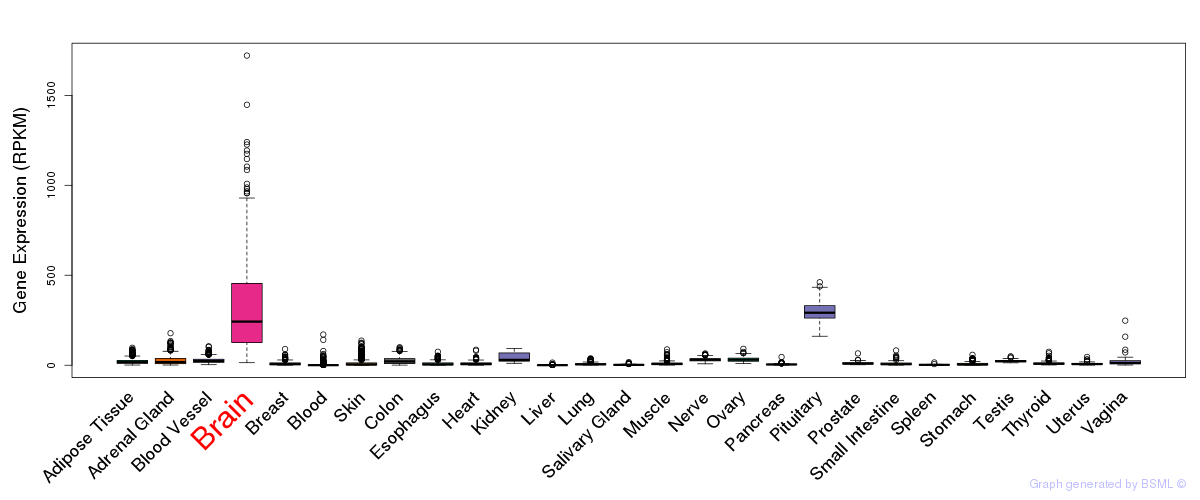
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFS5 | 0.91 | 0.88 |
| MRPS21 | 0.86 | 0.85 |
| NDUFA12 | 0.86 | 0.86 |
| NDUFA6 | 0.85 | 0.82 |
| PARK7 | 0.85 | 0.85 |
| MRPS18C | 0.84 | 0.82 |
| MRPL32 | 0.84 | 0.88 |
| NDUFS4 | 0.84 | 0.83 |
| ATP5O | 0.83 | 0.81 |
| TCEB1 | 0.83 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PEAR1 | -0.53 | -0.54 |
| TENC1 | -0.52 | -0.58 |
| FAM38A | -0.51 | -0.57 |
| LRRC32 | -0.49 | -0.54 |
| ENG | -0.49 | -0.50 |
| AF347015.26 | -0.48 | -0.42 |
| DOCK6 | -0.48 | -0.51 |
| TBX2 | -0.48 | -0.50 |
| MMRN2 | -0.48 | -0.47 |
| CASKIN2 | -0.47 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12082530 | |
| GO:0004197 | cysteine-type endopeptidase activity | IDA | 8639624 | |
| GO:0004221 | ubiquitin thiolesterase activity | TAS | 9521656 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008242 | omega peptidase activity | IDA | 9521656 | |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0043130 | ubiquitin binding | IDA | 9521656 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007412 | axon target recognition | IEA | axon (GO term level: 13) | - |
| GO:0019896 | axon transport of mitochondrion | IEA | axon (GO term level: 9) | - |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006950 | response to stress | IEA | - | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0007628 | adult walking behavior | IEA | - | |
| GO:0016579 | protein deubiquitination | IDA | 9521656 | |
| GO:0042755 | eating behavior | IEA | - | |
| GO:0050905 | neuromuscular process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0030424 | axon | IEA | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0005737 | cytoplasm | TAS | 16130169 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS UP | 44 | 23 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE UP | 85 | 57 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC DN | 59 | 35 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA DN | 11 | 6 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED EPIGENETICALLY IN PANCREATIC CANCER | 49 | 30 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG UP | 21 | 10 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MCGARVEY SILENCED BY METHYLATION IN COLON CANCER | 42 | 29 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D CLUSTER UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 DN | 39 | 21 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN DN | 27 | 15 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |