Gene Page: YWHAE
Summary ?
| GeneID | 7531 |
| Symbol | YWHAE |
| Synonyms | 14-3-3E|HEL2|KCIP-1|MDCR|MDS |
| Description | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| Reference | MIM:605066|HGNC:HGNC:12851|Ensembl:ENSG00000108953|HPRD:05457|Vega:OTTHUMG00000134316 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 1.016E-4 |
| Sherlock p-value | 0.241 |
| Fetal beta | 0.345 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.539 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
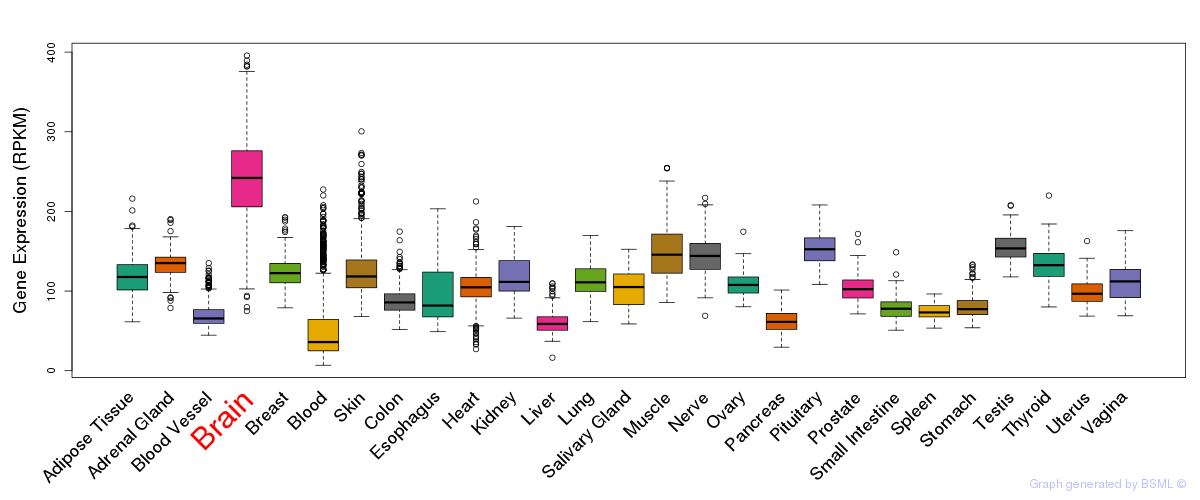
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF131 | 0.94 | 0.95 |
| DDX20 | 0.92 | 0.93 |
| ZNF615 | 0.92 | 0.93 |
| ZNF280D | 0.92 | 0.93 |
| ZNF567 | 0.91 | 0.92 |
| TOPORS | 0.91 | 0.92 |
| WDR43 | 0.91 | 0.92 |
| PHF20L1 | 0.91 | 0.92 |
| ZNF175 | 0.91 | 0.91 |
| C10orf137 | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.77 | -0.86 |
| AF347015.31 | -0.77 | -0.85 |
| FXYD1 | -0.76 | -0.86 |
| AF347015.33 | -0.75 | -0.83 |
| AF347015.27 | -0.75 | -0.84 |
| IFI27 | -0.75 | -0.85 |
| MT-CYB | -0.74 | -0.83 |
| AF347015.8 | -0.74 | -0.85 |
| HIGD1B | -0.74 | -0.86 |
| AIFM3 | -0.73 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019904 | protein domain specific binding | IEA | - | |
| GO:0019899 | enzyme binding | IPI | 10788521 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007242 | intracellular signaling cascade | TAS | 7644510 | |
| GO:0006605 | protein targeting | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | - | HPRD | 14970201 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD,BioGRID | 9369453 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | The cIAP-2 promoter interacts with 14-3-3-epsilon. | BIND | 15494311 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 10088721 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD,BioGRID | 7644510 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD | 7644510 |14559997 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | 14-3-3-epsilon interacts with cdc25A. | BIND | 7644510 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 7644510 |10713667 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | 14-3-3-epsilon interacts with cdc25B. | BIND | 7644510 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | Affinity Capture-Western | BioGRID | 10330186 |
| CDC2L1 | CDC2L2 | CDK11 | CDK11-p110 | CDK11-p46 | CDK11-p58 | CLK-1 | FLJ59152 | PK58 | p58 | p58CDC2L1 | p58CLK-1 | cell division cycle 2-like 1 (PITSLRE proteins) | CDK11-p110 interacts with 14-3-3-epsilon. | BIND | 15883043 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD | 12042314 |
| GPRIN2 | GRIN2 | KIAA0514 | MGC15171 | G protein regulated inducer of neurite outgrowth 2 | Two-hybrid | BioGRID | 16189514 |
| GRAP2 | GADS | GRAP-2 | GRB2L | GRBLG | GRID | GRPL | GrbX | Grf40 | Mona | P38 | GRB2-related adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10869435 |11504882 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | - | HPRD | 11486037 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | - | HPRD,BioGRID | 10869435 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 9111084 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | IGFIR interacts with 14-3-3-epsilon isoform. This interaction is modeled on a demonstrated interaction between human IGFIR and mouse 14-3-3-epsilon isoform | BIND | 9111084 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | IRS-1 interacts with 14-3-3-epsilon isoform. This interaction was modeled on a demonstrated interaction between human IRS-1 and mouse 14-3-3-epsilon isoform. | BIND | 9111084 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 9111084 |9312143 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 9312143 |
| KCNH2 | ERG1 | HERG | HERG1 | Kv11.1 | LQT2 | SQT1 | potassium voltage-gated channel, subfamily H (eag-related), member 2 | - | HPRD,BioGRID | 11953308 |
| KIF1C | KIAA0706 | LTXS1 | kinesin family member 1C | - | HPRD | 10559254 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 9524113 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Reconstituted Complex | BioGRID | 9452471 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | - | HPRD,BioGRID | 9427749 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | - | HPRD,BioGRID | 9452471 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD,BioGRID | 9452471 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 14743216 |
| MAPK7 | BMK1 | ERK4 | ERK5 | PRKM7 | mitogen-activated protein kinase 7 | Two-hybrid | BioGRID | 14679215 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | 14-3-3-epsilon interacts with SMRT. | BIND | 15494311 |
| NDEL1 | DKFZp451M0318 | EOPA | MITAP1 | NUDEL | nudE nuclear distribution gene E homolog (A. nidulans)-like 1 | - | HPRD,BioGRID | 12796778 |
| NGFRAP1 | BEX3 | Bex | DXS6984E | HGR74 | NADE | nerve growth factor receptor (TNFRSF16) associated protein 1 | - | HPRD,BioGRID | 11278287 |
| PAPOLA | MGC5378 | PAP | poly(A) polymerase alpha | - | HPRD | 14517258 |
| PCTK1 | FLJ16665 | PCTAIRE | PCTAIRE1 | PCTGAIRE | PCTAIRE protein kinase 1 | - | HPRD | 12154078 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 7644510 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | 14-3-3-epsilon interacts with Raf-1. | BIND | 7644510 |
| RASGRF1 | CDC25 | CDC25L | GNRP | GRF1 | GRF55 | H-GRF55 | PP13187 | Ras protein-specific guanine nucleotide-releasing factor 1 | - | HPRD | 11533041 |
| REM1 | GD:REM | GES | MGC48669 | REM | RAS (RAD and GEM)-like GTP-binding 1 | - | HPRD,BioGRID | 10441394 |
| RGNEF | DKFZp686P12164 | FLJ21817 | KIAA1998 | Rho-guanine nucleotide exchange factor | - | HPRD | 11533041 |
| RGS3 | C2PA | FLJ20370 | FLJ31516 | FLJ90496 | PDZ-RGS3 | RGP3 | regulator of G-protein signaling 3 | - | HPRD | 11985497 |
| RIN1 | - | Ras and Rab interactor 1 | - | HPRD | 9144171 |11784866 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 10407019 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | Two-hybrid | BioGRID | 16189514 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 8702721 |
| SYN2 | SYNII | SYNIIa | SYNIIb | synapsin II | Two-hybrid | BioGRID | 10358015 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD,BioGRID | 11172812 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 8702721 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | - | HPRD,BioGRID | 10788521 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Hamartin interacts with 14-3-3-epsilon. | BIND | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD | 12438239 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Tumor suppressor protein tuberin interacts with 14-3-3-epsilon isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-epsilon isoform. | BIND | 12176984 |
| WWTR1 | DKFZp586I1419 | TAZ | WW domain containing transcription regulator 1 | - | HPRD | 11118213 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY HIPPO | 22 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX2 DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 1 DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 2 UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-155 | 266 | 272 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-186 | 227 | 233 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-26 | 777 | 783 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-3p | 560 | 566 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC | ||||
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-31 | 608 | 614 | m8 | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-320 | 216 | 222 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-490 | 636 | 642 | 1A | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.