| KEGG AXON GUIDANCE
| 129 | 103 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN
| 182 | 111 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP
| 112 | 72 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN
| 104 | 59 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN
| 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN
| 133 | 77 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN
| 536 | 332 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP
| 176 | 123 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP
| 552 | 347 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP
| 97 | 60 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| SHI SPARC TARGETS DN
| 13 | 8 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP
| 414 | 287 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP
| 487 | 286 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP
| 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3
| 1118 | 744 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN
| 464 | 276 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN
| 108 | 84 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN
| 44 | 29 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL
| 50 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP
| 390 | 242 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN
| 830 | 547 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS DN
| 16 | 12 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP
| 262 | 186 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP
| 18 | 16 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN
| 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP
| 88 | 47 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1
| 528 | 324 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS
| 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC
| 210 | 123 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION
| 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN
| 195 | 135 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN
| 45 | 31 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN
| 154 | 101 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53
| 109 | 63 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP
| 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP
| 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN
| 701 | 446 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN
| 97 | 53 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN
| 546 | 362 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN
| 59 | 44 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN
| 264 | 159 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN
| 457 | 302 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 15
| 35 | 23 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS DN
| 32 | 27 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP
| 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS
| 62 | 35 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN
| 106 | 77 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 2HR
| 8 | 5 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR
| 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR
| 128 | 73 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED
| 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED
| 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME
| 1028 | 559 | All SZGR 2.0 genes in this pathway |
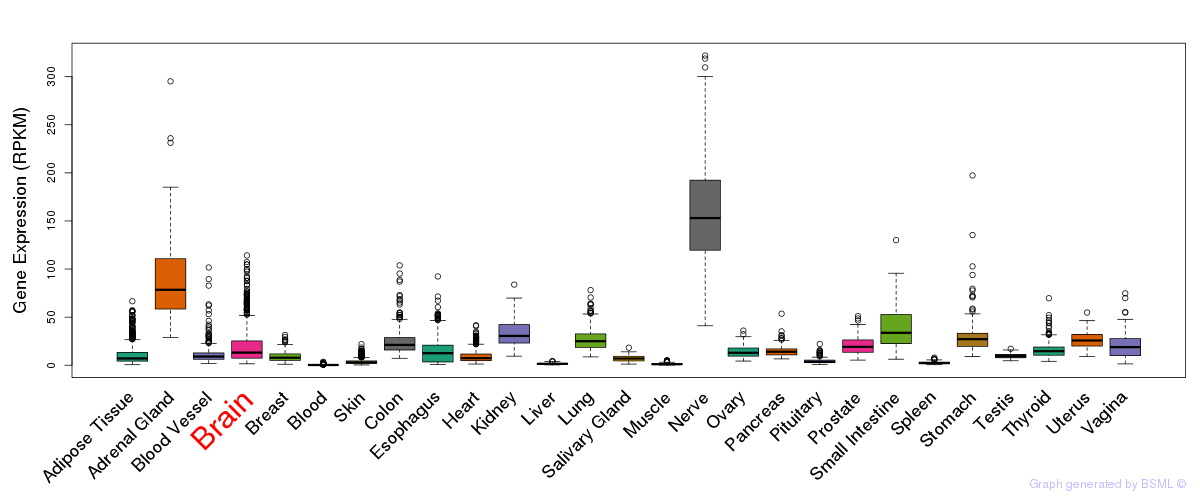
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation