Gene Page: GEMIN6
Summary ?
| GeneID | 79833 |
| Symbol | GEMIN6 |
| Synonyms | - |
| Description | gem nuclear organelle associated protein 6 |
| Reference | MIM:607006|HGNC:HGNC:20044|HPRD:06110| |
| Gene type | protein-coding |
| Map location | 2p22.1 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.915 |
| DMG | 1 (# studies) |
| eGene | Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07959896 | 2 | 39005451 | GEMIN6 | -0.027 | 0.3 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
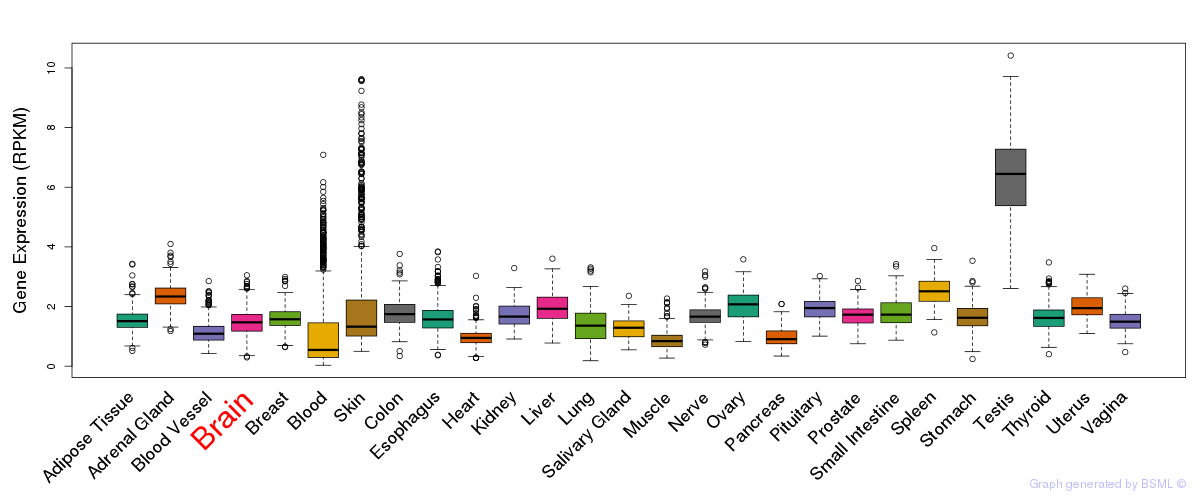
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | Affinity Capture-Western | BioGRID | 11748230 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-Western | BioGRID | 11748230 |
| GEMIN7 | SIP3 | gem (nuclear organelle) associated protein 7 | - | HPRD,BioGRID | 12065586 |
| SIP1 | GEMIN2 | SIP1-delta | survival of motor neuron protein interacting protein 1 | Affinity Capture-Western | BioGRID | 11748230 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | Affinity Capture-Western | BioGRID | 11748230 |
| SNRPB | COD | SNRPB1 | SmB/SmB' | snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 | Gemin6 interacts with SmB. | BIND | 11748230 |
| SNRPB | COD | SNRPB1 | SmB/SmB' | snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 | Gemin6 interacts with SNRPB (SmB) | BIND | 15939020 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | Gemin6 interacts with SmD2 | BIND | 11748230 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | Gemin6 interacts with SNRPD2 (SmD2) | BIND | 15939020 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | - | HPRD | 11748230 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | Gemin6 interacts with SNRPD3 (SmD3) | BIND | 15939020 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | Gemin6 interacts with SmD3. | BIND | 11748230 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | Gemin6 interacts with SmE. | BIND | 11748230 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | - | HPRD | 11748230 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | Gemin6 interacts with SNRPE (SmE) | BIND | 15939020 |
| SNRPF | SMF | small nuclear ribonucleoprotein polypeptide F | Gemin6 interacts with SNRPF (SmF) | BIND | 15939020 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | Gemin6 interacts with SNRPG (SmG) | BIND | 15939020 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE DN | 53 | 25 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |