Gene Page: GNPAT
Summary ?
| GeneID | 8443 |
| Symbol | GNPAT |
| Synonyms | DAP-AT|DAPAT|DHAPAT|RCDP2 |
| Description | glyceronephosphate O-acyltransferase |
| Reference | MIM:602744|HGNC:HGNC:4416|Ensembl:ENSG00000116906|HPRD:04120|Vega:OTTHUMG00000038024 |
| Gene type | protein-coding |
| Map location | 1q42 |
| Sherlock p-value | 0.163 |
| Fetal beta | 0.467 |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
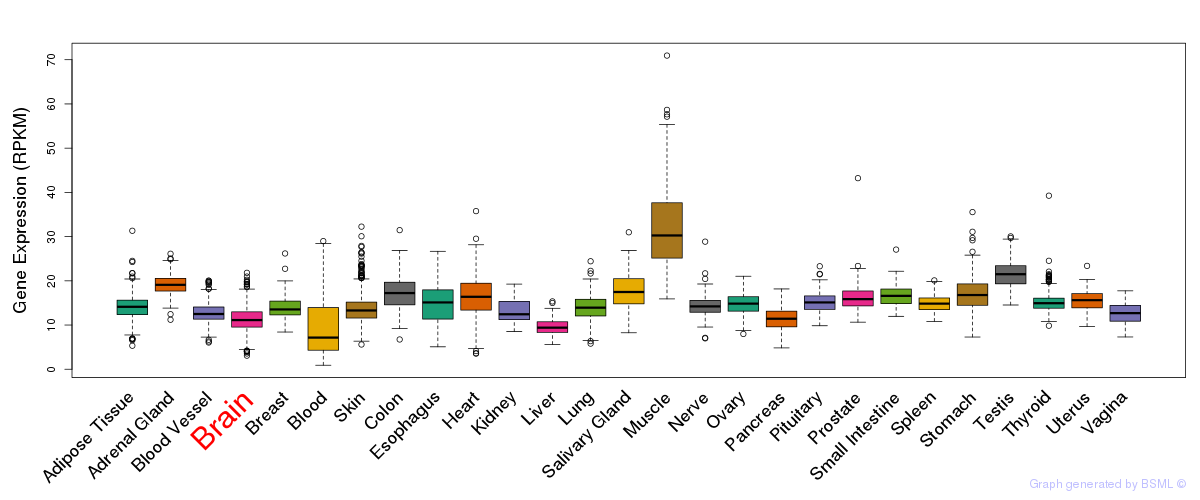
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C17orf68 | 0.94 | 0.95 |
| HMGXB3 | 0.92 | 0.94 |
| WDR59 | 0.92 | 0.93 |
| LZTR1 | 0.92 | 0.93 |
| INPP5E | 0.92 | 0.94 |
| INTS3 | 0.91 | 0.92 |
| FAM160B2 | 0.90 | 0.91 |
| HPS4 | 0.90 | 0.92 |
| DEPDC5 | 0.90 | 0.91 |
| GGA3 | 0.90 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.77 | -0.77 |
| AF347015.21 | -0.75 | -0.84 |
| MT-CO2 | -0.75 | -0.76 |
| AF347015.27 | -0.73 | -0.75 |
| MT-CYB | -0.72 | -0.72 |
| AF347015.8 | -0.71 | -0.74 |
| AF347015.33 | -0.70 | -0.70 |
| HIGD1B | -0.69 | -0.73 |
| AF347015.2 | -0.67 | -0.70 |
| AF347015.15 | -0.67 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008415 | acyltransferase activity | IEA | - | |
| GO:0008415 | acyltransferase activity | TAS | 9459311 | |
| GO:0016287 | glycerone-phosphate O-acyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009887 | organ morphogenesis | TAS | 9536089 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006631 | fatty acid metabolic process | TAS | 9459311 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10215861 | |
| GO:0005777 | peroxisome | TAS | 9459311 | |
| GO:0005778 | peroxisomal membrane | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCEROPHOSPHOLIPID METABOLISM | 77 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PA | 27 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | 82 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME PEROXISOMAL LIPID METABOLISM | 21 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |