Gene Page: RUNX2
Summary ?
| GeneID | 860 |
| Symbol | RUNX2 |
| Synonyms | AML3|CBF-alpha-1|CBFA1|CCD|CCD1|CLCD|OSF-2|OSF2|PEA2aA|PEBP2aA |
| Description | runt related transcription factor 2 |
| Reference | MIM:600211|HGNC:HGNC:10472|Ensembl:ENSG00000124813|HPRD:02566|Vega:OTTHUMG00000014774 |
| Gene type | protein-coding |
| Map location | 6p21 |
| Pascal p-value | 0.724 |
| Fetal beta | 0.237 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23376821 | 6 | 45391365 | RUNX2 | 7.01E-8 | -0.011 | 1.7E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
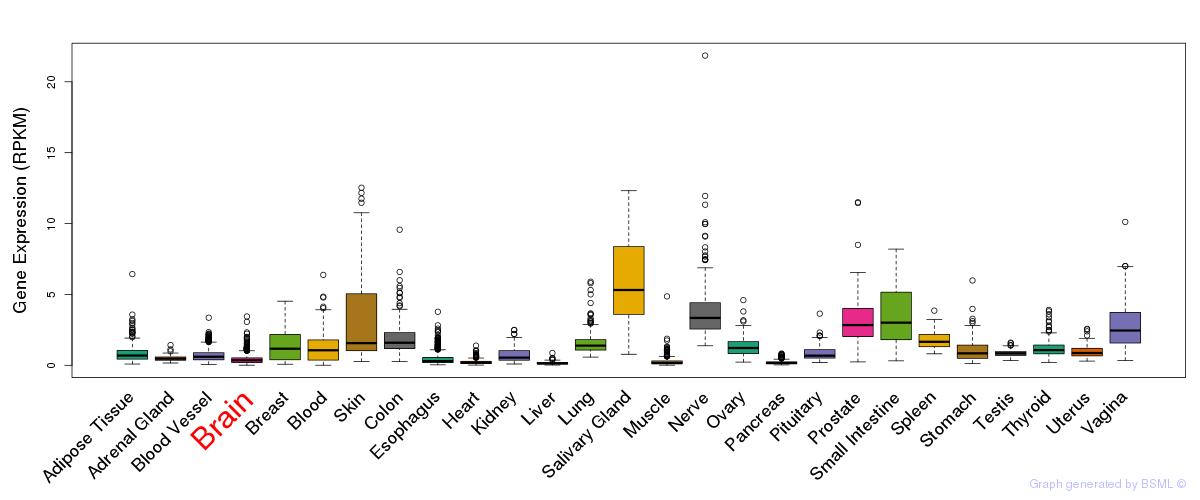
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBX2 | 0.68 | 0.63 |
| ITPKB | 0.67 | 0.57 |
| CHDH | 0.66 | 0.54 |
| TPCN1 | 0.65 | 0.56 |
| GPR37L1 | 0.65 | 0.56 |
| FAM38A | 0.64 | 0.58 |
| NACC2 | 0.64 | 0.56 |
| GPER | 0.64 | 0.60 |
| PBXIP1 | 0.64 | 0.56 |
| NFATC1 | 0.64 | 0.58 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.49 | -0.52 |
| C18orf21 | -0.47 | -0.49 |
| POLB | -0.46 | -0.52 |
| FRG1 | -0.46 | -0.52 |
| COQ3 | -0.46 | -0.48 |
| ING4 | -0.46 | -0.49 |
| PAK1IP1 | -0.45 | -0.46 |
| FARSB | -0.45 | -0.47 |
| ASNSD1 | -0.45 | -0.46 |
| DUSP12 | -0.45 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | NAS | 9182765 | |
| GO:0003702 | RNA polymerase II transcription factor activity | TAS | 9182762 | |
| GO:0005515 | protein binding | IPI | 11965546 |12145306 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016563 | transcription activator activity | IDA | 11965546 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0001649 | osteoblast differentiation | IEA | - | |
| GO:0001649 | osteoblast differentiation | TAS | 12217689 | |
| GO:0002062 | chondrocyte differentiation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0048469 | cell maturation | IEA | - | |
| GO:0016481 | negative regulation of transcription | IDA | 11965546 | |
| GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | IEA | - | |
| GO:0042487 | regulation of odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0045879 | negative regulation of smoothened signaling pathway | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CBFB | PEBP2B | core-binding factor, beta subunit | - | HPRD | 12434156 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD | 11668178 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD,BioGRID | 9794229 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD,BioGRID | 11641401 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15292260 |
| HDAC6 | FLJ16239 | HD6 | JM21 | histone deacetylase 6 | - | HPRD,BioGRID | 12391164 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11274169 |11641401 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | Reconstituted Complex | BioGRID | 12551949 |
| MSX2 | CRS2 | FPP | HOX8 | MSH | PFM | PFM1 | msh homeobox 2 | - | HPRD,BioGRID | 11683913 |
| MYST4 | DKFZp313G1618 | FLJ90335 | KAT6B | KIAA0383 | MORF | MOZ2 | qkf | querkopf | MYST histone acetyltransferase (monocytic leukemia) 4 | - | HPRD,BioGRID | 11965546 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Runx2 interacts with pRb. | BIND | 15583032 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western | BioGRID | 10531362 |10962029 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 10531362 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 10962029 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | Affinity Capture-Western | BioGRID | 10531362 |
| THOC4 | ALY | BEF | THO complex 4 | Reconstituted Complex | BioGRID | 9119228 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 10228168 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS | 14 | 12 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 1 | 46 | 33 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 1529 | 1535 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 1529 | 1535 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-130/301 | 2728 | 2734 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-135 | 1015 | 1021 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-137 | 1036 | 1042 | m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-148/152 | 2729 | 2735 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 741 | 747 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-19 | 2727 | 2733 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-203.1 | 934 | 940 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 3653 | 3659 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-205 | 1124 | 1131 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-218 | 1308 | 1315 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-221/222 | 3512 | 3518 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-224 | 623 | 629 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-23 | 1061 | 1067 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-5p | 3348 | 3354 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 1061 | 1067 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-324-5p | 2492 | 2498 | m8 | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-330 | 391 | 398 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-338 | 1242 | 1249 | 1A,m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-369-3p | 768 | 774 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 768 | 774 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-383 | 639 | 645 | 1A | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-448 | 741 | 747 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-485-3p | 1121 | 1128 | 1A,m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-495 | 3771 | 3777 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-543 | 2510 | 2516 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-9 | 2764 | 2770 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.