Gene Page: ACTN1
Summary ?
| GeneID | 87 |
| Symbol | ACTN1 |
| Synonyms | BDPLT15 |
| Description | actinin alpha 1 |
| Reference | MIM:102575|HGNC:HGNC:163|Ensembl:ENSG00000072110|HPRD:00020|Vega:OTTHUMG00000171386 |
| Gene type | protein-coding |
| Map location | 14q24|14q22-q24 |
| Pascal p-value | 0.387 |
| Sherlock p-value | 0.099 |
| TADA p-value | 0.018 |
| Fetal beta | -1.662 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 0 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACTN1 | chr14 | 69352217 | T | C | NM_001102 NM_001130004 NM_001130005 | p.437H>R p.437H>R p.437H>R | missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21350283 | 14 | 69445743 | ACTN1 | -0.024 | 0.27 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16827037 | chr3 | 117203284 | ACTN1 | 87 | 0.08 | trans |
Section II. Transcriptome annotation
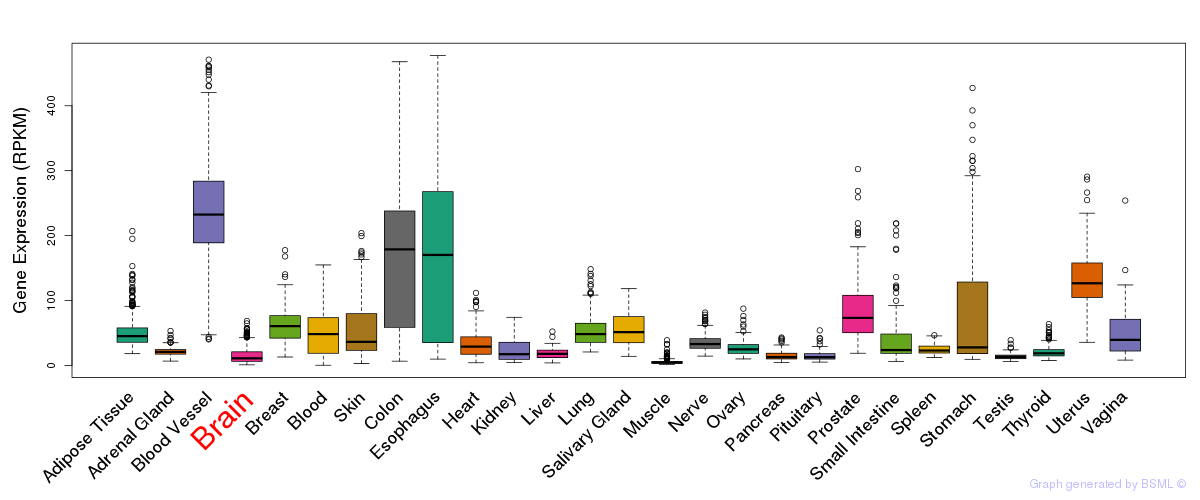
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDC2L5 | 0.87 | 0.90 |
| ZBTB2 | 0.87 | 0.88 |
| KLHL20 | 0.86 | 0.91 |
| SART3 | 0.86 | 0.89 |
| UBTF | 0.86 | 0.88 |
| KIAA0174 | 0.86 | 0.88 |
| RSPRY1 | 0.85 | 0.89 |
| ZNF146 | 0.85 | 0.88 |
| CTR9 | 0.85 | 0.88 |
| SMEK1 | 0.85 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.71 | -0.81 |
| AF347015.31 | -0.70 | -0.80 |
| MT-CO2 | -0.70 | -0.80 |
| AF347015.33 | -0.68 | -0.77 |
| AF347015.27 | -0.68 | -0.78 |
| IFI27 | -0.68 | -0.77 |
| C5orf53 | -0.67 | -0.71 |
| AF347015.21 | -0.66 | -0.83 |
| HIGD1B | -0.66 | -0.78 |
| MT-CYB | -0.66 | -0.76 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | - | HPRD,BioGRID | 11470434 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western | BioGRID | 9535896 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12223541 |
| CDK5R2 | NCK5AI | P39 | p39nck5ai | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | - | HPRD,BioGRID | 12223541 |
| COL17A1 | BA16H23.2 | BP180 | BPAG2 | FLJ60881 | KIAA0204 | LAD-1 | collagen, type XVII, alpha 1 | - | HPRD,BioGRID | 11739652 |
| CSRP3 | CLP | CMD1M | CMH12 | CRP3 | LMO4 | MGC14488 | MGC61993 | MLP | cysteine and glycine-rich protein 3 (cardiac LIM protein) | - | HPRD | 12642359 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | - | HPRD,BioGRID | 7790378 |
| FBP1 | FBP | fructose-1,6-bisphosphatase 1 | - | HPRD,BioGRID | 12507293 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10198040 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 11854440 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | Reconstituted Complex | BioGRID | 2116421 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MYOT | LGMD1 | LGMD1A | TTID | myotilin | - | HPRD,BioGRID | 12499399 |
| MYOZ1 | CS-2 | FATZ | MYOZ | myozenin 1 | - | HPRD | 11842093 |
| MYOZ1 | CS-2 | FATZ | MYOZ | myozenin 1 | Two-hybrid | BioGRID | 16189514 |
| MYOZ2 | C4orf5 | CS-1 | myozenin 2 | - | HPRD | 11842093 |
| MYOZ3 | CS-3 | CS3 | FRP3 | myozenin 3 | - | HPRD | 11842093 |
| NEBL | FLJ53769 | LNEBL | MGC119746 | MGC119747 | bA56H7.1 | nebulette | - | HPRD | 10470015 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | - | HPRD,BioGRID | 11110697 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | CLP-36 interacts with alpha-actinin 1. This interaction was modeled on a demonstrated interaction between human CLP-36 and chicken alpha-actinin 1. | BIND | 10753915 |
| PHB | PHB1 | prohibitin | Affinity Capture-Western | BioGRID | 12628297 |
| PKN1 | DBK | MGC46204 | PAK1 | PKN | PKN-ALPHA | PRK1 | PRKCL1 | protein kinase N1 | - | HPRD,BioGRID | 9030526 |11802708 |
| PYGB | MGC9213 | phosphorylase, glycogen; brain | Reconstituted Complex | BioGRID | 12211109 |
| RAVER1 | KIAA1978 | ribonucleoprotein, PTB-binding 1 | - | HPRD | 11724819 |
| SSX2IP | ADIP | FLJ10848 | KIAA0923 | MGC75026 | synovial sarcoma, X breakpoint 2 interacting protein | - | HPRD,BioGRID | 12446711 |
| TES | DKFZp586B2022 | MGC1146 | TESS | TESS-2 | testis derived transcript (3 LIM domains) | - | HPRD | 12695497 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | TRAF4 interacts with alpha-actinin. | BIND | 12023963 |
| TTN | CMD1G | CMH9 | CMPD4 | CONNECTIN | DKFZp451N061 | EOMFC | FLJ26020 | FLJ26409 | FLJ32040 | FLJ34413 | FLJ39564 | FLJ43066 | HMERF | LGMD2J | TMD | titin | - | HPRD,BioGRID | 11305911 |11846417 |
| XRCC4 | - | X-ray repair complementing defective repair in Chinese hamster cells 4 | Two-hybrid | BioGRID | 16189514 |
| ZYX | ESP-2 | HED-2 | zyxin | - | HPRD,BioGRID | 10224105 |11423549 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELL2CELL PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| LANG MYB FAMILY TARGETS | 29 | 16 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS DN | 28 | 18 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |