Gene Page: SUCLA2
Summary ?
| GeneID | 8803 |
| Symbol | SUCLA2 |
| Synonyms | A-BETA|MTDPS5|SCS-betaA |
| Description | succinate-CoA ligase, ADP-forming, beta subunit |
| Reference | MIM:603921|HGNC:HGNC:11448|Ensembl:ENSG00000136143|HPRD:06798|Vega:OTTHUMG00000016889 |
| Gene type | protein-coding |
| Map location | 13q14.2 |
| Pascal p-value | 0.818 |
| Sherlock p-value | 0.685 |
| Fetal beta | -0.666 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_mitochondria G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08280787 | 13 | 48575775 | SUCLA2 | 1.3E-8 | -0.011 | 5.2E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1570589 | chr9 | 116874354 | SUCLA2 | 8803 | 0.17 | trans |
Section II. Transcriptome annotation
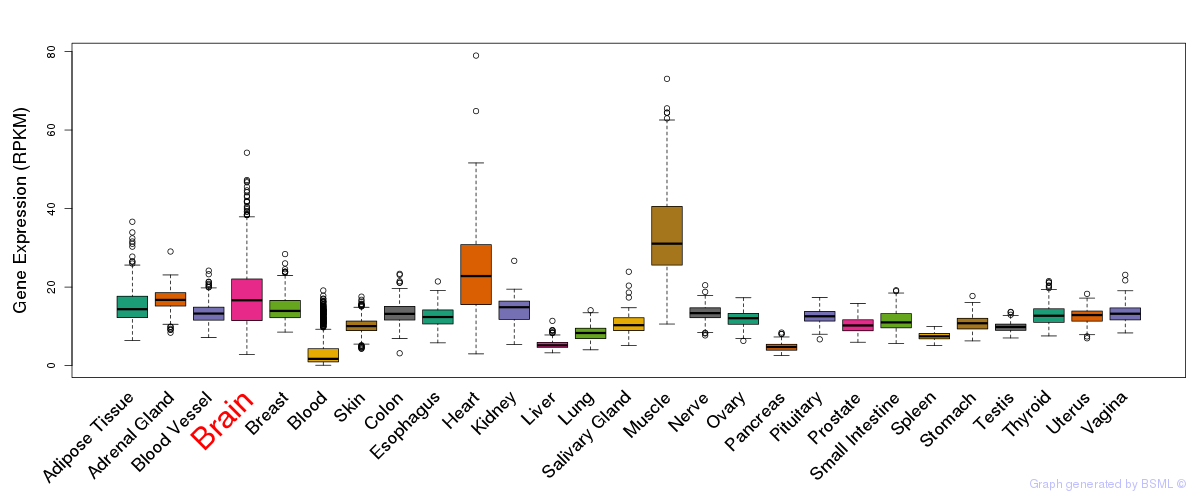
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBM4 | 0.95 | 0.94 |
| POLR3D | 0.95 | 0.96 |
| MKRN1 | 0.95 | 0.96 |
| EIF4A1 | 0.95 | 0.94 |
| IP6K2 | 0.95 | 0.93 |
| GPN1 | 0.95 | 0.95 |
| NIPSNAP1 | 0.95 | 0.95 |
| PGD | 0.94 | 0.93 |
| PRPF6 | 0.94 | 0.92 |
| C14orf133 | 0.94 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.91 |
| MT-CO2 | -0.78 | -0.91 |
| AF347015.27 | -0.78 | -0.91 |
| AF347015.33 | -0.78 | -0.91 |
| AIFM3 | -0.76 | -0.83 |
| MT-CYB | -0.76 | -0.90 |
| FXYD1 | -0.76 | -0.90 |
| AF347015.8 | -0.75 | -0.91 |
| TSC22D4 | -0.75 | -0.84 |
| HLA-F | -0.75 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CITRATE CYCLE TCA CYCLE | 32 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG PROPANOATE METABOLISM | 33 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KREB PATHWAY | 8 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | 48 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | 141 | 85 | All SZGR 2.0 genes in this pathway |
| REACTOME CITRIC ACID CYCLE TCA CYCLE | 26 | 16 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MOOTHA TCA | 16 | 13 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |