Gene Page: HAP1
Summary ?
| GeneID | 9001 |
| Symbol | HAP1 |
| Synonyms | HAP2|HIP5|HLP|hHLP1 |
| Description | huntingtin-associated protein 1 |
| Reference | MIM:600947|HGNC:HGNC:4812|Ensembl:ENSG00000173805|HPRD:02972|Vega:OTTHUMG00000133498 |
| Gene type | protein-coding |
| Map location | 17q21.2-q21.3 |
| Pascal p-value | 0.674 |
| Sherlock p-value | 0.128 |
| Fetal beta | -0.604 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0229 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
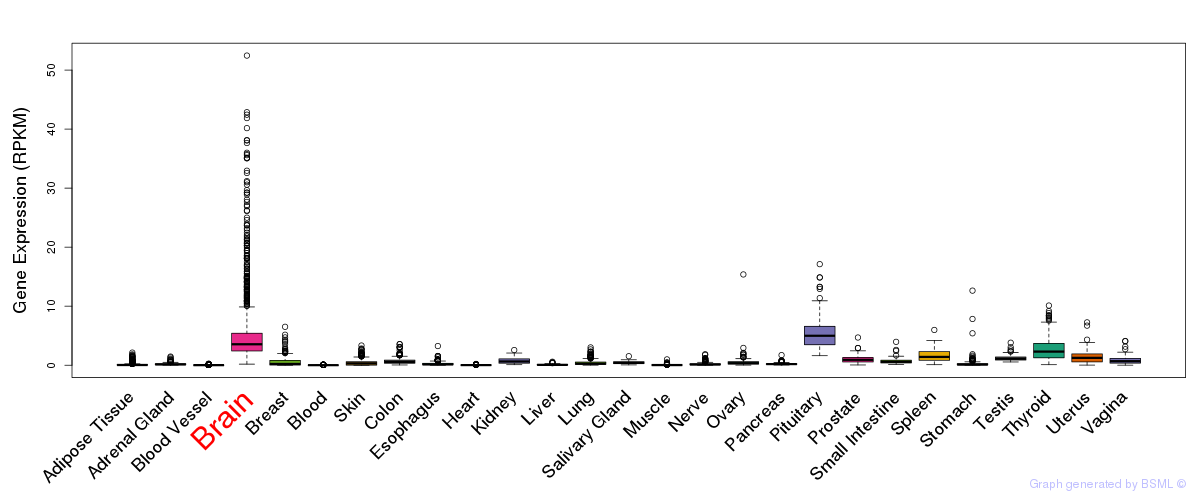
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRIG3 | 0.78 | 0.31 |
| COL4A6 | 0.77 | 0.50 |
| BOC | 0.77 | 0.35 |
| SOX3 | 0.75 | 0.32 |
| NHLH1 | 0.75 | 0.40 |
| MFAP2 | 0.74 | 0.43 |
| TNFRSF19 | 0.74 | 0.47 |
| VIM | 0.74 | 0.42 |
| GPC4 | 0.73 | 0.38 |
| JUB | 0.72 | 0.35 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.33 | -0.34 |
| AF347015.27 | -0.31 | -0.37 |
| LGI4 | -0.30 | -0.23 |
| AF347015.33 | -0.30 | -0.36 |
| AC131097.1 | -0.30 | -0.38 |
| AF347015.31 | -0.29 | -0.33 |
| HLA-F | -0.29 | -0.26 |
| AF347015.8 | -0.29 | -0.35 |
| MT-CO2 | -0.29 | -0.35 |
| AIFM3 | -0.28 | -0.25 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9361024 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0015629 | actin cytoskeleton | TAS | 9361024 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP5J2 | ATP5JL | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | Two-hybrid | BioGRID | 16169070 |
| BRD7 | BP75 | CELTIX1 | NAG4 | bromodomain containing 7 | - | HPRD | 15383276 |
| C15orf29 | FLJ22557 | MGC117356 | chromosome 15 open reading frame 29 | Two-hybrid | BioGRID | 16169070 |
| C8orf33 | FLJ20989 | chromosome 8 open reading frame 33 | Two-hybrid | BioGRID | 16169070 |
| CBX8 | HPC3 | PC3 | RC1 | chromobox homolog 8 (Pc class homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| CCDC113 | DKFZp434N1418 | FLJ23555 | HSPC065 | coiled-coil domain containing 113 | Two-hybrid | BioGRID | 16169070 |
| CDK5RAP2 | C48 | Cep215 | DKFZp686B1070 | DKFZp686D1070 | KIAA1633 | MCPH3 | CDK5 regulatory subunit associated protein 2 | Two-hybrid | BioGRID | 16169070 |
| COL9A2 | DJ39G22.4 | EDM2 | MED | collagen, type IX, alpha 2 | Two-hybrid | BioGRID | 16169070 |
| CRIP1 | CRHP | CRIP | CRP1 | FLJ40971 | cysteine-rich protein 1 (intestinal) | Two-hybrid | BioGRID | 16169070 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD,BioGRID | 9361024 |9454836 |
| DDX49 | FLJ10432 | R27090_2 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 | Two-hybrid | BioGRID | 16169070 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | Two-hybrid | BioGRID | 16169070 |
| FAM173A | C16orf24 | MGC2494 | family with sequence similarity 173, member A | Two-hybrid | BioGRID | 16169070 |
| GLTSCR2 | PICT-1 | PICT1 | glioma tumor suppressor candidate region gene 2 | Two-hybrid | BioGRID | 16169070 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | - | HPRD,BioGRID | 9798945 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD,BioGRID | 12021262 |
| HMOX2 | HO-2 | heme oxygenase (decycling) 2 | Two-hybrid | BioGRID | 16169070 |
| HTT | HD | IT15 | huntingtin | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 7477378 |9454836 |9599014 |9668110 |
| HTT | HD | IT15 | huntingtin | Htt interacts with HAP1. This interaction was modeled on a demonstrated interaction between human Htt and Rat HAP1. | BIND | 15654337 |
| HTT | HD | IT15 | huntingtin | Huntingtin interacts with HAP1. | BIND | 9668110 |
| HTT | HD | IT15 | huntingtin | Htt interacts with Hap1. | BIND | 7477378 |
| HTT | HD | IT15 | huntingtin | - | HPRD | 7477378 |9599014 |9668110 |10974549 |
| MPP3 | DLG3 | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | Two-hybrid | BioGRID | 16169070 |
| MRPS9 | MRP-S9 | RPMS9 | S9mt | mitochondrial ribosomal protein S9 | Two-hybrid | BioGRID | 16169070 |
| NAP1L5 | DRLM | nucleosome assembly protein 1-like 5 | Two-hybrid | BioGRID | 16169070 |
| NDUFB9 | B22 | DKFZp566O173 | FLJ22885 | LYRM3 | UQOR22 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa | Two-hybrid | BioGRID | 16169070 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | - | HPRD,BioGRID | 12881483 |
| NIPSNAP3A | DKFZp564D177 | FLJ13953 | HSPC299 | MGC14553 | NIPSNAP4 | TASSC | nipsnap homolog 3A (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| PCM1 | PTC4 | pericentriolar material 1 | - | HPRD,BioGRID | 9361024 |
| PDCD7 | ES18 | HES18 | MGC22015 | programmed cell death 7 | Two-hybrid | BioGRID | 16169070 |
| PFDN1 | PDF | PFD1 | prefoldin subunit 1 | Two-hybrid | BioGRID | 16169070 |
| PPID | CYP-40 | CYPD | MGC33096 | peptidylprolyl isomerase D | Two-hybrid | BioGRID | 16169070 |
| PPOX | MGC8485 | PPO | V290M | VP | protoporphyrinogen oxidase | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| RER1 | - | RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| RPS10 | MGC88819 | ribosomal protein S10 | Two-hybrid | BioGRID | 16169070 |
| RPS25 | - | ribosomal protein S25 | Two-hybrid | BioGRID | 16169070 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | Two-hybrid | BioGRID | 16169070 |
| SNAPIN | SNAPAP | SNAP-associated protein | Two-hybrid | BioGRID | 16169070 |
| STX5 | SED5 | STX5A | syntaxin 5 | Two-hybrid | BioGRID | 16169070 |
| TAF1D | JOSD3 | MGC5306 | TAF(I)41 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa | Two-hybrid | BioGRID | 16169070 |
| TNNT3 | AMCD2B | DA2B | DKFZp779M2348 | FSSV | troponin T type 3 (skeletal, fast) | Two-hybrid | BioGRID | 16169070 |
| TOMM20 | KIAA0016 | MAS20 | MGC117367 | MOM19 | TOM20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | Two-hybrid | BioGRID | 16169070 |
| TSPYL1 | TSPYL | TSPY-like 1 | Two-hybrid | BioGRID | 16169070 |
| UTP3 | CRL1 | CRLZ1 | FLJ23256 | SAS10 | UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| ZNF20 | FLJ39241 | KOX13 | zinc finger protein 20 | Two-hybrid | BioGRID | 16169070 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
| ZNF691 | RP11-342M1.5 | Zfp691 | zinc finger protein 691 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| ASGHARZADEH NEUROBLASTOMA POOR SURVIVAL DN | 46 | 30 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE GENES | 39 | 20 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-328 | 405 | 411 | m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-339 | 402 | 408 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.