Gene Page: SERTAD2
Summary ?
| GeneID | 9792 |
| Symbol | SERTAD2 |
| Synonyms | Sei-2|TRIP-Br2 |
| Description | SERTA domain containing 2 |
| Reference | HGNC:HGNC:30784|Ensembl:ENSG00000179833|HPRD:18040|Vega:OTTHUMG00000152678 |
| Gene type | protein-coding |
| Map location | 2p14 |
| Pascal p-value | 0.209 |
| Sherlock p-value | 0.03 |
| Fetal beta | 0.775 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06836181 | 2 | 64995545 | SERTAD2 | 1.69E-10 | -0.022 | 5.18E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17126801 | chr1 | 65098162 | SERTAD2 | 9792 | 0.11 | trans | ||
| snp_a-1990354 | 0 | SERTAD2 | 9792 | 0.2 | trans | |||
| rs6707117 | chr2 | 136855504 | SERTAD2 | 9792 | 0.04 | trans | ||
| rs16839796 | chr4 | 7188348 | SERTAD2 | 9792 | 0.02 | trans | ||
| rs10040715 | chr5 | 57594207 | SERTAD2 | 9792 | 0.06 | trans | ||
| rs10945911 | chr6 | 164047509 | SERTAD2 | 9792 | 0.13 | trans | ||
| rs1419793 | chr7 | 34697630 | SERTAD2 | 9792 | 0.2 | trans | ||
| rs16911034 | chr11 | 23302320 | SERTAD2 | 9792 | 0.13 | trans | ||
| rs2460047 | chr11 | 93678935 | SERTAD2 | 9792 | 0.12 | trans | ||
| rs1885073 | chr14 | 104357181 | SERTAD2 | 9792 | 0.06 | trans |
Section II. Transcriptome annotation
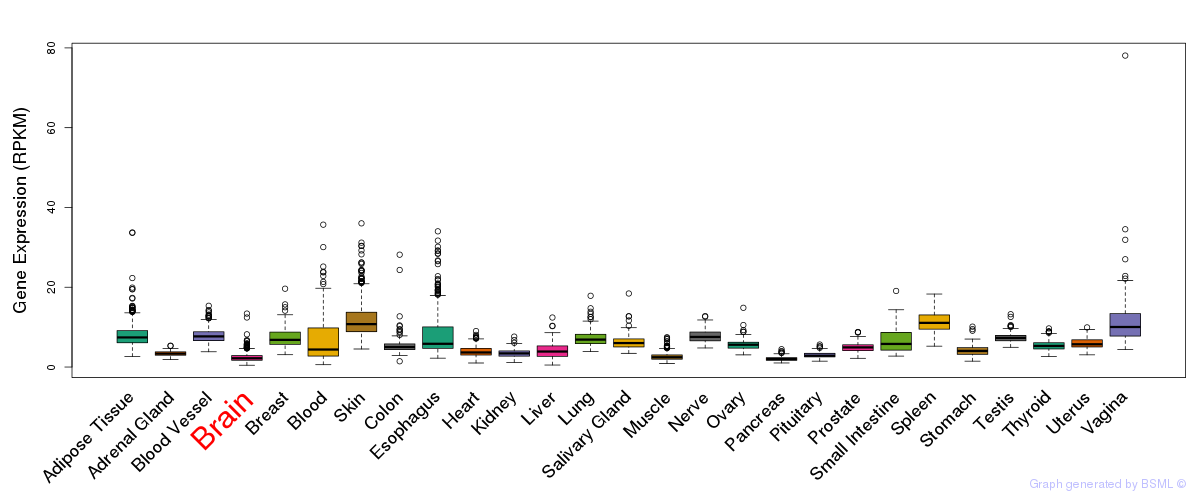
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LY75 | 0.93 | 0.95 |
| PON2 | 0.81 | 0.79 |
| ADD3 | 0.80 | 0.74 |
| HRSP12 | 0.80 | 0.77 |
| RANBP3L | 0.79 | 0.83 |
| TLR3 | 0.78 | 0.76 |
| GPC5 | 0.77 | 0.82 |
| SDC2 | 0.77 | 0.77 |
| NT5E | 0.76 | 0.80 |
| PLSCR4 | 0.75 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF821 | -0.58 | -0.66 |
| MPP3 | -0.57 | -0.65 |
| KIAA1543 | -0.57 | -0.64 |
| PODXL2 | -0.57 | -0.65 |
| DBN1 | -0.56 | -0.63 |
| FAM40A | -0.56 | -0.63 |
| ANKRD13B | -0.56 | -0.64 |
| CSNK1E | -0.56 | -0.62 |
| DPF1 | -0.56 | -0.64 |
| AXIN1 | -0.56 | -0.63 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| PACHER TARGETS OF IGF1 AND IGF2 UP | 35 | 27 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS UP | 48 | 36 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| GENTILE RESPONSE CLUSTER D3 | 61 | 39 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |