Gene Page: MED24
Summary ?
| GeneID | 9862 |
| Symbol | MED24 |
| Synonyms | ARC100|CRSP100|CRSP4|DRIP100|MED5|THRAP4|TRAP100 |
| Description | mediator complex subunit 24 |
| Reference | MIM:607000|HGNC:HGNC:22963|Ensembl:ENSG00000008838|HPRD:06107|Vega:OTTHUMG00000133329 |
| Gene type | protein-coding |
| Map location | 17q21.1 |
| Pascal p-value | 0.035 |
| Sherlock p-value | 0.72 |
| Fetal beta | -0.177 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17740645 | 17 | 37894413 | MED24 | 0.001 | 2.577 | DMG:vanEijk_2014 | |
| cg13878456 | 17 | 38020344 | MED24 | 3.332E-4 | -4.089 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11220437 | chr11 | 126148159 | MED24 | 9862 | 0.18 | trans | ||
| rs8177388 | chr11 | 126167473 | MED24 | 9862 | 0.05 | trans |
Section II. Transcriptome annotation
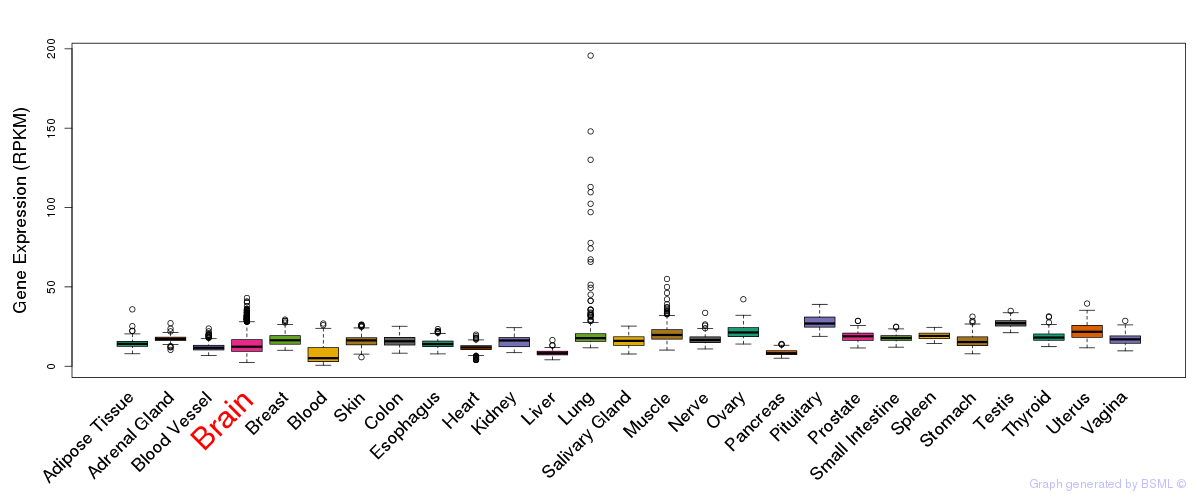
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB18 | 0.87 | 0.85 |
| RAB5A | 0.84 | 0.81 |
| DLAT | 0.84 | 0.81 |
| PTPLAD1 | 0.84 | 0.83 |
| PPP2CA | 0.84 | 0.81 |
| COPS8 | 0.83 | 0.81 |
| NBN | 0.82 | 0.83 |
| UBLCP1 | 0.82 | 0.80 |
| STX12 | 0.82 | 0.80 |
| ACTR2 | 0.82 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.61 | -0.43 |
| MT-CO2 | -0.60 | -0.51 |
| AF347015.18 | -0.57 | -0.47 |
| HIGD1B | -0.57 | -0.47 |
| AF347015.2 | -0.56 | -0.47 |
| AF347015.31 | -0.55 | -0.48 |
| MT-CYB | -0.55 | -0.47 |
| AF347015.8 | -0.55 | -0.46 |
| CSAG1 | -0.54 | -0.44 |
| IL32 | -0.53 | -0.35 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15208681 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-MS | BioGRID | 10198638 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 9653119 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-MS | BioGRID | 12837248 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Co-purification | BioGRID | 9653119 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS | BioGRID | 15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD | 14576168 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| PPARA | MGC2237 | MGC2452 | NR1C1 | PPAR | hPPAR | peroxisome proliferator-activated receptor alpha | - | HPRD,BioGRID | 9653119 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD,BioGRID | 9653119 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 9653119 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 10198638 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-MS | BioGRID | 10198638 |12837248 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 6 | 75 | 48 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |