Gene Page: CTR9
Summary ?
| GeneID | 9646 |
| Symbol | CTR9 |
| Synonyms | SH2BP1|TSBP|p150|p150TSP |
| Description | CTR9 homolog, Paf1/RNA polymerase II complex component |
| Reference | MIM:609366|HGNC:HGNC:16850|Ensembl:ENSG00000198730|HPRD:06717|Vega:OTTHUMG00000165789 |
| Gene type | protein-coding |
| Map location | 11p15.3 |
| Pascal p-value | 0.877 |
| Sherlock p-value | 0.207 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4971079 | chr1 | 155130390 | CTR9 | 9646 | 0.09 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
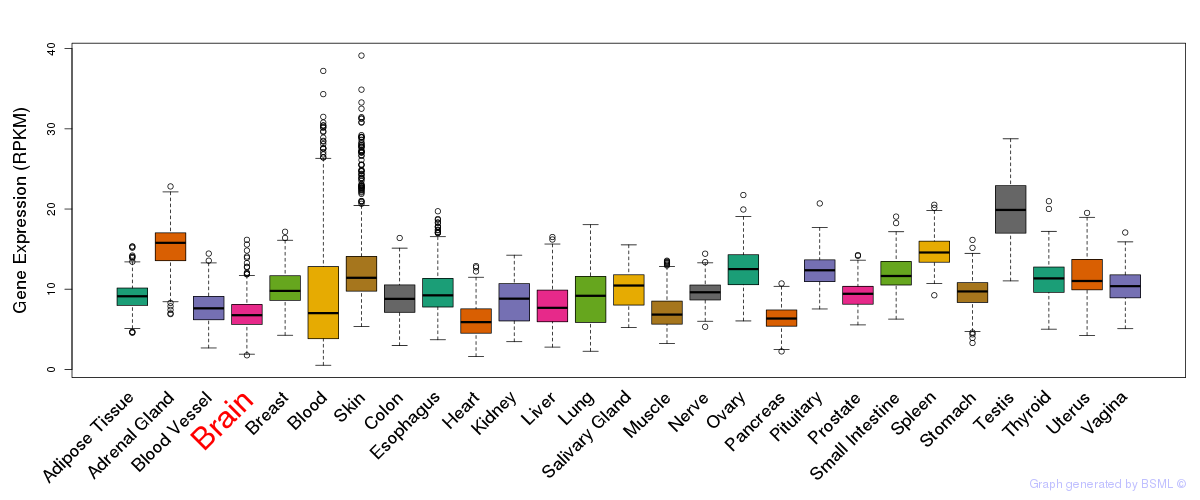
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RFC1 | 0.92 | 0.92 |
| DDX42 | 0.92 | 0.92 |
| FKBP15 | 0.90 | 0.89 |
| GIGYF2 | 0.89 | 0.90 |
| ZNF638 | 0.89 | 0.91 |
| HNRNPUL2 | 0.89 | 0.89 |
| SUPT6H | 0.89 | 0.89 |
| ARID4B | 0.89 | 0.91 |
| SLTM | 0.89 | 0.91 |
| TPR | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.75 | -0.77 |
| AF347015.21 | -0.74 | -0.77 |
| IFI27 | -0.73 | -0.75 |
| HIGD1B | -0.72 | -0.75 |
| MT-CO2 | -0.72 | -0.74 |
| ENHO | -0.71 | -0.77 |
| FXYD1 | -0.70 | -0.72 |
| C1orf54 | -0.69 | -0.75 |
| VAMP5 | -0.68 | -0.71 |
| MYL3 | -0.68 | -0.71 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |