Gene Page: CDKN1B
Summary ?
| GeneID | 1027 |
| Symbol | CDKN1B |
| Synonyms | CDKN4|KIP1|MEN1B|MEN4|P27KIP1 |
| Description | cyclin-dependent kinase inhibitor 1B |
| Reference | MIM:600778|HGNC:HGNC:1785|Ensembl:ENSG00000111276|HPRD:02867|Vega:OTTHUMG00000149914 |
| Gene type | protein-coding |
| Map location | 12p13.1-p12 |
| Pascal p-value | 0.249 |
| Sherlock p-value | 8.849E-6 |
| Fetal beta | 0.697 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0925 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
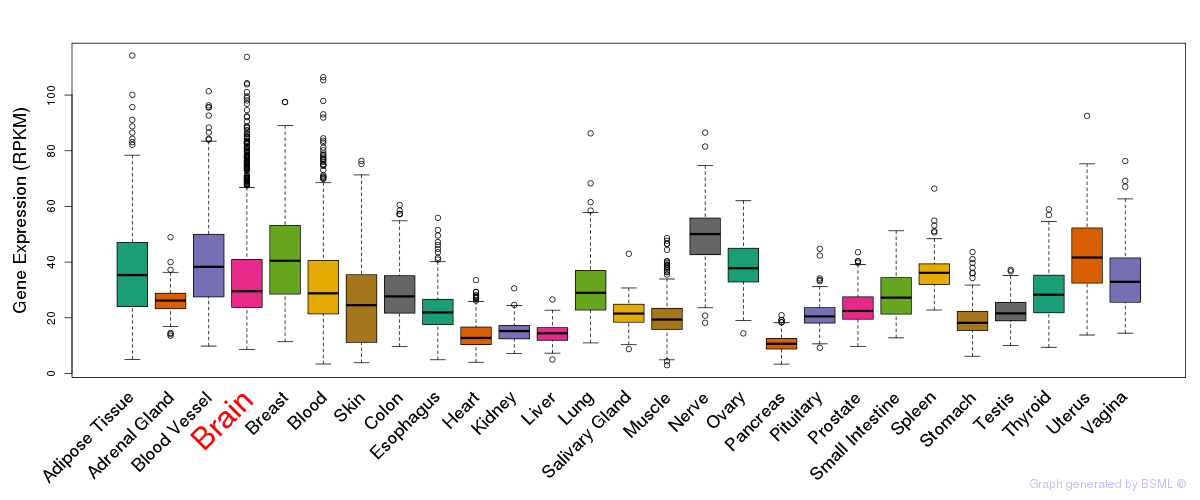
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAD2L1 | 0.90 | 0.72 |
| CCNB1 | 0.90 | 0.72 |
| CENPN | 0.89 | 0.74 |
| CKS2 | 0.87 | 0.59 |
| CHEK2 | 0.87 | 0.65 |
| PBK | 0.86 | 0.32 |
| KIAA0101 | 0.86 | 0.31 |
| HMGB2 | 0.86 | 0.55 |
| RAD51AP1 | 0.86 | 0.46 |
| AURKA | 0.86 | 0.52 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.34 | -0.28 |
| AF347015.15 | -0.33 | -0.26 |
| AF347015.8 | -0.33 | -0.25 |
| MT-CYB | -0.33 | -0.26 |
| APOL1 | -0.32 | -0.27 |
| AF347015.2 | -0.32 | -0.24 |
| AF347015.33 | -0.31 | -0.24 |
| MT-CO2 | -0.31 | -0.22 |
| SLC9A3R2 | -0.31 | -0.23 |
| AF347015.18 | -0.31 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity | TAS | 8033212 | |
| GO:0005515 | protein binding | IPI | 8756624 |9106657 |9840943 |12082530 |15057270 |15652749 |15890360 |17254966 |17254967 | |
| GO:0004861 | cyclin-dependent protein kinase inhibitor activity | TAS | 10918569 | |
| GO:0004860 | protein kinase inhibitor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000079 | regulation of cyclin-dependent protein kinase activity | TAS | 8033212 | |
| GO:0000082 | G1/S transition of mitotic cell cycle | IDA | 10208428 | |
| GO:0006917 | induction of apoptosis | IDA | 10208428 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007050 | cell cycle arrest | IMP | 12093740 | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 8033212 | |
| GO:0048102 | autophagic cell death | IDA | 12698196 | |
| GO:0042326 | negative regulation of phosphorylation | IDA | 10208428 | |
| GO:0030308 | negative regulation of cell growth | IDA | 10208428 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 12244303 | |
| GO:0005634 | nucleus | IDA | 12093740 | |
| GO:0005737 | cytoplasm | IDA | 12093740 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 12042314 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Two-hybrid | BioGRID | 16169070 |
| CCNA1 | - | cyclin A1 | cyclin A1 interacts with p27. | BIND | 10022926 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | p27 interacts with Cyc A. | BIND | 15469821 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | p27 interacts with cyclin A. | BIND | 9632134 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD,BioGRID | 12501191 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | p27 interacts with cyclin A. | BIND | 15652749 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | in vitro Reconstituted Complex | BioGRID | 10908655 |11360184 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | p27 interacts with cyclin D. | BIND | 9632134 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD | 7478582 |10908655 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | Two-hybrid | BioGRID | 16189514 |
| CCND3 | - | cyclin D3 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10342870 |11360184 |16189514 |
| CCNE1 | CCNE | cyclin E1 | p27 interacts with cyclin E. | BIND | 9632134 |
| CCNE1 | CCNE | cyclin E1 | p27 interacts with Cyc E. | BIND | 15469821 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 9891079 |12529437 |
| CCNE2 | CYCE2 | cyclin E2 | - | HPRD | 9858585 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | p27 interacts with an unspecified isoform of CDK2. | BIND | 15652749 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Cdk2 interacts with p27kip1. | BIND | 15619620 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD,BioGRID | 12972555 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Cdk2 interacts with p27. | BIND | 15469821 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD,BioGRID | 10908655 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | Affinity Capture-Western | BioGRID | 11360184 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | - | HPRD | 9858585 |
| CKS1B | CKS1 | PNAS-16 | PNAS-18 | ckshs1 | CDC28 protein kinase regulatory subunit 1B | Biochemical Activity Reconstituted Complex | BioGRID | 12140288 |12813041 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | Affinity Capture-Western | BioGRID | 12082530 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | COPS5 (CSN5) interacts with CDKN1B (p27). | BIND | 15688030 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | FOXO3a interacts with p27Kip1 promoter. | BIND | 15084260 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 11278754 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | p27 interacts with importin-alpha. | BIND | 15579463 |
| NUP50 | MGC39961 | NPAP60 | NPAP60L | nucleoporin 50kDa | - | HPRD,BioGRID | 10891500 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | - | HPRD | 10559916 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 12042314 |12140288 |12813041 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | CDKN1B (p27Kip1) interacts with SKP2 (p45Skp2). | BIND | 15735731 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | Skp2 interacts with p27. | BIND | 15469821 |
| SPDYA | MGC110856 | MGC57218 | Ringo3 | SPDY1 | SPY1 | speedy homolog A (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12972555 |
| STMN1 | LAP18 | Lag | OP18 | PP17 | PP19 | PR22 | SMN | stathmin 1/oncoprotein 18 | p27 interacts with stathmin. | BIND | 15652749 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Affinity Capture-Western | BioGRID | 15355997 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western | BioGRID | 15355997 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | Ub interacts with p27. | BIND | 15469821 |
| UCHL1 | PARK5 | PGP9.5 | Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | - | HPRD,BioGRID | 12082530 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | p27Kip1 interacts with CRM1. | BIND | 11889117 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD,BioGRID | 11889117 |12529437 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD | 12042314 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 12042314 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD,BioGRID | 12042314 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCM PATHWAY | 18 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P27 PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SA REG CASCADE OF CYCLIN EXPR | 13 | 7 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | 56 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 5 UP | 16 | 7 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN | 49 | 29 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA | 47 | 35 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| SARTIPY BLUNTED BY INSULIN RESISTANCE DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| MATZUK LUTEAL GENES | 8 | 5 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 MULTIPLE MYELOMA PROGRAM | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SCHOEN NFKB SIGNALING | 34 | 26 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-148/152 | 128 | 134 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-186 | 427 | 433 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-190 | 90 | 96 | m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-194 | 1301 | 1307 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-196 | 245 | 251 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-221/222 | 201 | 208 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC | ||||
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-24 | 1253 | 1260 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-323 | 1275 | 1281 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-377 | 1278 | 1285 | 1A,m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-455 | 279 | 285 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.