Gene Page: CDKN2A
Summary ?
| GeneID | 1029 |
| Symbol | CDKN2A |
| Synonyms | ARF|CDK4I|CDKN2|CMM2|INK4|INK4A|MLM|MTS-1|MTS1|P14|P14ARF|P16|P16-INK4A|P16INK4|P16INK4A|P19|P19ARF|TP16 |
| Description | cyclin-dependent kinase inhibitor 2A |
| Reference | MIM:600160|HGNC:HGNC:1787|Ensembl:ENSG00000147889|HPRD:02542|Vega:OTTHUMG00000019686 |
| Gene type | protein-coding |
| Map location | 9p21 |
| Pascal p-value | 0.032 |
| Fetal beta | -0.468 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0889 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00718440 | 9 | 21995312 | CDKN2A | -0.026 | 0.35 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | CDKN2A | 1029 | 0.18 | trans |
Section II. Transcriptome annotation
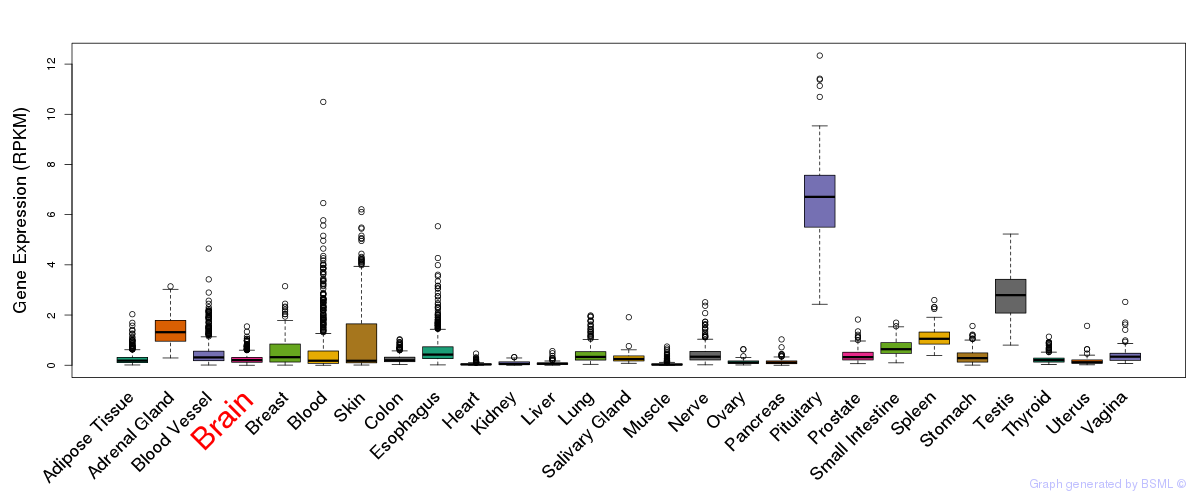
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11278317 |12740913 |15989956 |17274640 |17909018 | |
| GO:0004861 | cyclin-dependent protein kinase inhibitor activity | IDA | 8259215 | |
| GO:0051059 | NF-kappaB binding | IDA | 10353611 | |
| GO:0019901 | protein kinase binding | IPI | 8259215 | |
| GO:0055105 | ubiquitin-protein ligase inhibitor activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000075 | cell cycle checkpoint | IMP | 16243918 | |
| GO:0000082 | G1/S transition of mitotic cell cycle | IDA | 10208428 | |
| GO:0001953 | negative regulation of cell-matrix adhesion | IMP | 10205165 | |
| GO:0006309 | DNA fragmentation during apoptosis | IMP | 12082630 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006469 | negative regulation of protein kinase activity | IMP | 15582998 | |
| GO:0006364 | rRNA processing | IEA | - | |
| GO:0006917 | induction of apoptosis | IDA | 10208428 | |
| GO:0006917 | induction of apoptosis | IMP | 12082630 | |
| GO:0007050 | cell cycle arrest | IMP | 15582998 |16243918 | |
| GO:0008285 | negative regulation of cell proliferation | IMP | 16243918 | |
| GO:0008637 | apoptotic mitochondrial changes | IMP | 12082630 | |
| GO:0010149 | senescence | IMP | 14720514 |14966292 | |
| GO:0048103 | somatic stem cell division | ISS | - | |
| GO:0010389 | regulation of G2/M transition of mitotic cell cycle | IMP | 15582998 | |
| GO:0006919 | caspase activation | IMP | 12082630 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0042326 | negative regulation of phosphorylation | IDA | 8259215 |10208428 | |
| GO:0033088 | negative regulation of immature T cell proliferation in the thymus | ISS | - | |
| GO:0032088 | negative regulation of NF-kappaB transcription factor activity | IDA | 10353611 | |
| GO:0030308 | negative regulation of cell growth | IDA | 10208428 | |
| GO:0031647 | regulation of protein stability | ISS | - | |
| GO:0030889 | negative regulation of B cell proliferation | ISS | - | |
| GO:0045736 | negative regulation of cyclin-dependent protein kinase activity | IDA | 8259215 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0051444 | negative regulation of ubiquitin-protein ligase activity | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 16243918 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IDA | 17110379 | |
| GO:0005730 | nucleolus | IDA | 17110379 | |
| GO:0005737 | cytoplasm | IDA | 16243918 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALS2CR8 | CARF | DKFZp667N246 | FLJ12675 | FLJ21579 | NYD-SP24 | amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8 | - | HPRD | 12154087 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | Affinity Capture-MS | BioGRID | 17353931 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Reconstituted Complex | BioGRID | 10580009 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | Reconstituted Complex | BioGRID | 15065884 |
| CCNG1 | CCNG | cyclin G1 | - | HPRD,BioGRID | 12556559 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | p16 interacts with cdk4. This interaction was modeled on a demonstrated interaction between human p16 and cdk4 from an unspecified species. | BIND | 9482104 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | CDKN2A interacts with CDK4 | BIND | 11556834 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD,BioGRID | 9228064 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD | 9228064 |15232106 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | p16 interacts with CDK4. This interaction was modeled on a demonstrated interaction between human p16 and CDK4 from an unspecified species. | BIND | 10491434 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | p16 interacts with cdk4. | BIND | 8805225 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | CDKN2A interacts with CDK6 | BIND | 11556834 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | p16 interacts with CDK6. This interaction was modeled on a demonstrated interaction between human p16 and CDK6 from an unspecified species. | BIND | 10491434 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | p16 interacts with cdk6. This interaction was modeled on a demonstrated interaction between human p16 and cdk6 from an unspecified species. | BIND | 9482104 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | p16 interacts with cdk6. | BIND | 8805225 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | - | HPRD,BioGRID | 9751050 |
| CDKN2AIP | CARF | FLJ20036 | CDKN2A interacting protein | - | HPRD | 12154087 |
| CDKN2C | INK4C | p18 | p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | - | HPRD | 8259215 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | p14ARF interacts with E2F-1. | BIND | 12082609 |
| E4F1 | E4F | MGC99614 | E4F transcription factor 1 | - | HPRD,BioGRID | 12446718 |
| FKBP2 | FKBP-13 | PPIase | FK506 binding protein 2, 13kDa | Affinity Capture-Western | BioGRID | 12606707 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | p14ARF interacts with HIF-1alpha. | BIND | 11382768 |
| HUWE1 | ARF-BP1 | HECTH9 | HSPC272 | Ib772 | KIAA0312 | LASU1 | MULE | UREB1 | HECT, UBA and WWE domain containing 1 | ARF interacts with ARF-BP1. | BIND | 15989956 |
| MAPK10 | FLJ12099 | FLJ33785 | JNK3 | JNK3A | MGC50974 | PRKM10 | p493F12 | p54bSAPK | mitogen-activated protein kinase 10 | CDKN2A (p16INK4a) interacts with MAPK10 (JNK3) | BIND | 16007099 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | CDKN2A (p16INK4a) interacts with MAPK8 (JNK1) | BIND | 16007099 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD | 15232106 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 9529248 |12085228|9529249 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p14ARF interacts with an unspecified isoform of Mdm2. | BIND | 12582152 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9529248 |9529249 |12085228 |14612427 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with p14ARF. | BIND | 10822382 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p14 interacts with MDM2. | BIND | 15355988 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | MDM2 interacts with p14ARF | BIND | 9653180 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | Affinity Capture-Western | BioGRID | 11297540 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | p14ARF interacts with c-Myc. | BIND | 15361884 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | NCOR1 (N-CoR) interacts with p-p14(ARF) (p14(ARF) promoter). | BIND | 15729358 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | NCOR2 (SMRT) interacts with p-p14(ARF) (p14(ARF) promoter). | BIND | 15729358 |
| PLEKHA8 | FAPP2 | MGC3358 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 | Reconstituted Complex | BioGRID | 15107860 |
| PPP1R9B | FLJ30345 | PPP1R6 | PPP1R9 | SPINO | Spn | protein phosphatase 1, regulatory (inhibitor) subunit 9B | - | HPRD,BioGRID | 11278317 |
| PPP1R9B | FLJ30345 | PPP1R6 | PPP1R9 | SPINO | Spn | protein phosphatase 1, regulatory (inhibitor) subunit 9B | SPINO interacts with p14ARF. | BIND | 11278317 |
| PSMC3 | MGC8487 | TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | p14ARF interacts with TBP-1. | BIND | 14665636 |
| RPL11 | GIG34 | ribosomal protein L11 | Affinity Capture-Western | BioGRID | 14612427 |
| SERTAD1 | SEI1 | TRIP-Br1 | SERTA domain containing 1 | - | HPRD,BioGRID | 10580009 |15065884 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 9529249 |12446718 |14612427 |
| UBE2A | HHR6A | RAD6A | UBC2 | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | Affinity Capture-Western | BioGRID | 12640129 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | p14 interacts with WRN. | BIND | 15355988 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | YY1 interacts with p14ARF. | BIND | 15210108 |
| ZNF410 | APA-1 | APA1 | zinc finger protein 410 | - | HPRD | 12370286 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARF PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SA REG CASCADE OF CYCLIN EXPR | 13 | 7 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| SLEBOS HEAD AND NECK CANCER WITH HPV UP | 84 | 43 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS DN | 14 | 14 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| TAKADA GASTRIC CANCER COPY NUMBER DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| BUSA SAM68 TARGETS UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| HONRADO BREAST CANCER BRCA1 VS BRCA2 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| OHM EMBRYONIC CARCINOMA DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| SUZUKI RESPONSE TO TSA AND DECITABINE 1A | 23 | 16 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED EPIGENETICALLY IN PANCREATIC CANCER | 49 | 30 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PARTIALLY REPROGRAMMED TO PLURIPOTENCY | 10 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |