Gene Page: GNA13
Summary ?
| GeneID | 10672 |
| Symbol | GNA13 |
| Synonyms | G13 |
| Description | G protein subunit alpha 13 |
| Reference | MIM:604406|HGNC:HGNC:4381|HPRD:05100| |
| Gene type | protein-coding |
| Map location | 17q24.3 |
| Pascal p-value | 1.378E-4 |
| Sherlock p-value | 0.407 |
| Fetal beta | 1.28 |
| DMG | 1 (# studies) |
| Support | G-PROTEIN RELAY CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1115 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23423852 | 17 | 63053202 | GNA13 | 8.55E-9 | 0.005 | 3.98E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
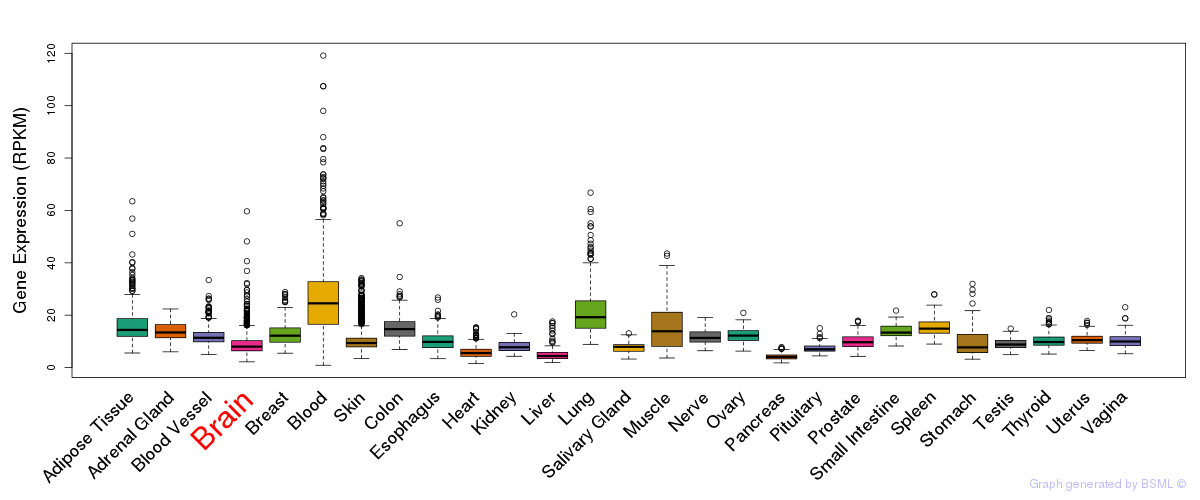
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 7791744 | |
| GO:0005515 | protein binding | IPI | 10026210 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0019001 | guanyl nucleotide binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001569 | patterning of blood vessels | IEA | - | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007266 | Rho protein signal transduction | IEA | - | |
| GO:0007243 | protein kinase cascade | IEA | - | |
| GO:0007165 | signal transduction | TAS | 7791744 | |
| GO:0030168 | platelet activation | IEA | - | |
| GO:0008360 | regulation of cell shape | IEA | - | |
| GO:0006928 | cell motion | TAS | 8999798 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030334 | regulation of cell migration | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP3 | AKAP110 | FSP95 | PRKA3 | SOB1 | A kinase (PRKA) anchor protein 3 | - | HPRD,BioGRID | 11696326 |
| ARHGEF1 | GEF1 | LBCL2 | LSC | P115-RHOGEF | SUB1.5 | Rho guanine nucleotide exchange factor (GEF) 1 | - | HPRD | 9641916 |
| ARHGEF1 | GEF1 | LBCL2 | LSC | P115-RHOGEF | SUB1.5 | Rho guanine nucleotide exchange factor (GEF) 1 | GNA13 (alpha-13) interacts with ARHGEF1 (p115RhoGEF). | BIND | 15735747 |
| ARHGEF1 | GEF1 | LBCL2 | LSC | P115-RHOGEF | SUB1.5 | Rho guanine nucleotide exchange factor (GEF) 1 | - | HPRD,BioGRID | 9641916 |12681510 |
| ARHGEF11 | DKFZp667F1223 | GTRAP48 | KIAA0380 | PDZ-RHOGEF | Rho guanine nucleotide exchange factor (GEF) 11 | - | HPRD,BioGRID | 10026210 |
| ARHGEF12 | DKFZp686O2372 | KIAA0382 | LARG | PRO2792 | Rho guanine nucleotide exchange factor (GEF) 12 | - | HPRD,BioGRID | 11094164 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD,BioGRID | 11136230 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | - | HPRD,BioGRID | 11136230 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 11136230 |
| DRD5 | DBDR | DRD1B | DRD1L2 | MGC10601 | dopamine receptor D5 | - | HPRD,BioGRID | 12623966 |
| EDNRB | ABCDS | ETB | ETBR | ETRB | HSCR | HSCR2 | endothelin receptor type B | - | HPRD | 12135322 |
| F2R | CF2R | HTR | PAR1 | TR | coagulation factor II (thrombin) receptor | Affinity Capture-Western | BioGRID | 8290554 |
| MCF2 | DBL | MCF.2 cell line derived transforming sequence | in vitro in vivo | BioGRID | 11062019 |
| MTNR1A | MEL-1A-R | MT1 | melatonin receptor 1A | - | HPRD | 10598579 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | GNA13 (alpha-13) interacts with PPP5C (PP5). | BIND | 15735747 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD,BioGRID | 12176367 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | Affinity Capture-Western | BioGRID | 11696326 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | - | HPRD,BioGRID | 8824244 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | - | HPRD,BioGRID | 8824244 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD,BioGRID | 8824244 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10821841 |
| RDX | DFNB24 | radixin | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10816569 |
| RGS16 | A28-RGS14 | A28-RGS14P | RGS-R | regulator of G-protein signaling 16 | - | HPRD,BioGRID | 14634662 |
| RIC8A | MGC104517 | MGC131931 | MGC148073 | MGC148074 | RIC8 | synembryn | resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | - | HPRD,BioGRID | 12509430 |
| S1PR2 | AGR16 | EDG-5 | EDG5 | Gpcr13 | H218 | LPB2 | S1P2 | sphingosine-1-phosphate receptor 2 | - | HPRD | 10488065 |
| S1PR3 | EDG-3 | EDG3 | FLJ37523 | FLJ93220 | LPB3 | MGC71696 | S1P3 | sphingosine-1-phosphate receptor 3 | - | HPRD | 10488065 |
| TBXA2R | TXA2-R | thromboxane A2 receptor | - | HPRD,BioGRID | 8290554|10395940 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | Phenotypic Enhancement | BioGRID | 12515866 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD | 8552586 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MYOSIN PATHWAY | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP13 PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P4 PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR4 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID S1P META PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL AMPLIFICATION | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP | 69 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL DN | 74 | 42 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS LIGHTYELLOW UP | 11 | 8 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE UP | 136 | 80 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| SU THYMUS | 20 | 12 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION DN | 13 | 7 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 2681 | 2687 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 2681 | 2687 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-30-5p | 615 | 621 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.