Gene Page: GIPC1
Summary ?
| GeneID | 10755 |
| Symbol | GIPC1 |
| Synonyms | C19orf3|GIPC|GLUT1CBP|Hs.6454|IIP-1|NIP|RGS19IP1|SEMCAP|SYNECTIIN|SYNECTIN|TIP-2 |
| Description | GIPC PDZ domain containing family member 1 |
| Reference | MIM:605072|HGNC:HGNC:1226|Ensembl:ENSG00000123159|HPRD:05462|Vega:OTTHUMG00000182248 |
| Gene type | protein-coding |
| Map location | 19p13.1 |
| Pascal p-value | 0.019 |
| Sherlock p-value | 0.731 |
| Fetal beta | -1.127 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.7595 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7729934 | chr5 | 136104636 | GIPC1 | 10755 | 0.18 | trans |
Section II. Transcriptome annotation
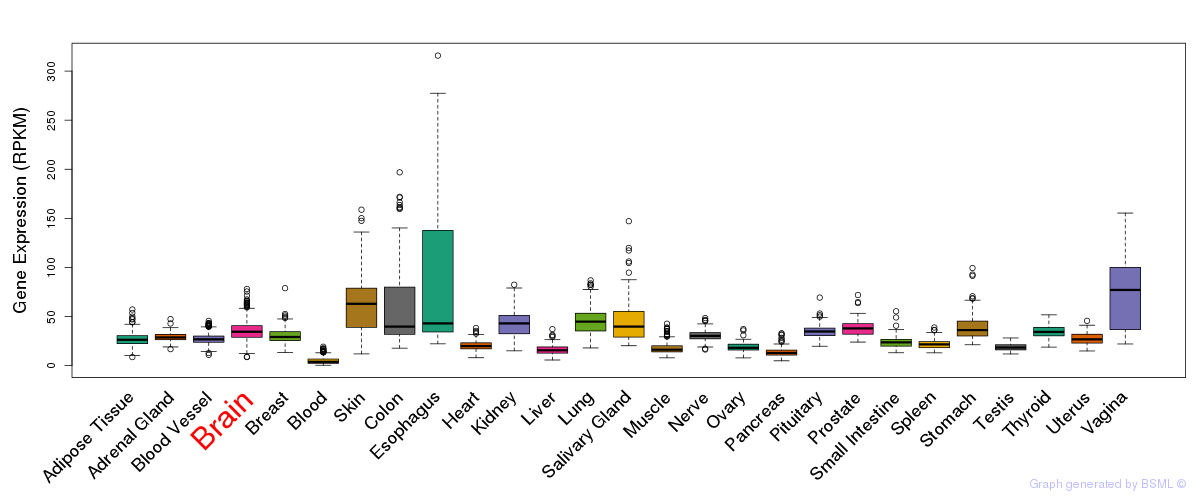
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IPI | Neurotransmitter (GO term level: 4) | 15459234 |
| GO:0003779 | actin binding | IEA | - | |
| GO:0017022 | myosin binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0014047 | glutamate secretion | IEA | glutamate, Neurotransmitter (GO term level: 7) | - |
| GO:0048167 | regulation of synaptic plasticity | IEA | Synap (GO term level: 8) | - |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | NAS | 9770488 | |
| GO:0006605 | protein targeting | IEA | - | |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | ISS | - | |
| GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | ISS | - | |
| GO:0031647 | regulation of protein stability | ISS | - | |
| GO:0043542 | endothelial cell migration | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IEA | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0043198 | dendritic shaft | IEA | dendrite (GO term level: 7) | - |
| GO:0043197 | dendritic spine | IEA | Synap, dendrite (GO term level: 7) | - |
| GO:0012506 | vesicle membrane | IEA | - | |
| GO:0005829 | cytosol | ISS | - | |
| GO:0005624 | membrane fraction | IDA | 9770488 | |
| GO:0005625 | soluble fraction | IDA | 9770488 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | ISS | - | |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | IDA | 12857860 | |
| GO:0005938 | cell cortex | IDA | 12857860 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10198040 |
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | Affinity Capture-Western Two-hybrid | BioGRID | 12724327 |
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | The carboxy terminus of beta-1-AR interacts with GIPC. | BIND | 12724327 |
| CD93 | C1QR1 | C1qR(P) | C1qRP | CDw93 | MXRA4 | dJ737E23.1 | CD93 molecule | CD93 interacts with GIPC. | BIND | 15459234 |
| CD93 | C1QR1 | C1qR(P) | C1qRP | CDw93 | MXRA4 | dJ737E23.1 | CD93 molecule | GIPC interacts with CD93. This interaction was modelled on a demonstrated interaction between GIPC from an unspecified species and human CD93. | BIND | 15819698 |
| DRD3 | D3DR | ETM1 | FET1 | MGC149204 | MGC149205 | dopamine receptor D3 | Two-hybrid | BioGRID | 14499480 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | - | HPRD,BioGRID | 11479315 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | - | HPRD,BioGRID | 11479315 |
| KIF1B | CMT2 | CMT2A | CMT2A1 | FLJ23699 | HMSNII | KIAA0591 | KIAA1448 | KLP | MGC134844 | kinesin family member 1B | - | HPRD,BioGRID | 10198040 |
| LHCGR | FLJ41504 | LCGR | LGR2 | LH/CG-R | LH/CGR | LHR | LHRHR | LSH-R | luteinizing hormone/choriogonadotropin receptor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14507927 |
| LHCGR | FLJ41504 | LCGR | LGR2 | LH/CG-R | LH/CGR | LHR | LHRHR | LSH-R | luteinizing hormone/choriogonadotropin receptor | GIPC interacts with hLHR. | BIND | 14507927 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | Reconstituted Complex Two-hybrid | BioGRID | 10827173 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD,BioGRID | 11912251 |
| LRP8 | APOER2 | HSZ75190 | MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | Two-hybrid | BioGRID | 10827173 |
| MARS | FLJ35667 | METRS | MTRNS | methionyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Affinity Capture-MS | BioGRID | 17353931 |
| MMS19 | FLJ34167 | FLJ95146 | MET18 | MGC99604 | MMS19L | hMMS19 | MMS19 nucleotide excision repair homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| MYO1C | FLJ23903 | MMI-beta | MMIb | NMI | myr2 | myosin IC | Affinity Capture-MS | BioGRID | 17353931 |
| MYO6 | DFNA22 | DFNB37 | KIAA0389 | myosin VI | - | HPRD | 12857860 |12893809 |
| MYO6 | DFNA22 | DFNB37 | KIAA0389 | myosin VI | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12857860 |17353931 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | GIPC interacts with TRKA. This interaction was modelled on a demonstrated interaction between mouse GIPC and rat TrkA. | BIND | 11251075 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 11251075 |
| NTRK2 | GP145-TrkB | TRKB | neurotrophic tyrosine kinase, receptor, type 2 | - | HPRD,BioGRID | 11251075 |
| NUP93 | KIAA0095 | MGC21106 | nucleoporin 93kDa | Affinity Capture-MS | BioGRID | 17353931 |
| PDXDC1 | KIAA0251 | LP8165 | pyridoxal-dependent decarboxylase domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RGS1 | 1R20 | BL34 | IER1 | IR20 | regulator of G-protein signaling 1 | - | HPRD | 11251075 |
| RGS19 | GAIP | RGSGAIP | regulator of G-protein signaling 19 | GAIP interacts with GIPC. This interaction was modelled on a demonstrated interaction between human GAIP and mouse GIPC. | BIND | 11251075 |
| RGS19 | GAIP | RGSGAIP | regulator of G-protein signaling 19 | - | HPRD,BioGRID | 9770488 |11251075 |11956658 |
| SDC4 | MGC22217 | SYND4 | syndecan 4 | Affinity Capture-Western | BioGRID | 10911369 |
| SEMA4C | FLJ20369 | KIAA1739 | M-SEMA-F | MGC126382 | MGC126383 | SEMACL1 | SEMAF | SEMAI | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | in vitro in vivo | BioGRID | 10318831 |
| SH3BP4 | BOG25 | TTP | SH3-domain binding protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | - | HPRD,BioGRID | 10198040 |
| TAGLN | DKFZp686P11128 | SM22 | SMCC | TAGLN1 | WS3-10 | transgelin | Affinity Capture-MS | BioGRID | 17353931 |
| TPBG | 5T4 | 5T4-AG | M6P1 | trophoblast glycoprotein | - | HPRD,BioGRID | 11798178 |
| TYRP1 | CAS2 | CATB | GP75 | TRP | TYRP | b-PROTEIN | tyrosinase-related protein 1 | - | HPRD,BioGRID | 11441007 |
| ZFPM1 | FOG | FOG1 | ZNF408 | ZNF89A | zinc finger protein, multitype 1 | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 3 DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 230 | 237 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-194 | 481 | 487 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-491 | 223 | 229 | m8 | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.