Gene Page: YWHAQ
Summary ?
| GeneID | 10971 |
| Symbol | YWHAQ |
| Synonyms | 14-3-3|1C5|HS1 |
| Description | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| Reference | MIM:609009|HGNC:HGNC:12854|Ensembl:ENSG00000134308|HPRD:00886|Vega:OTTHUMG00000013848 |
| Gene type | protein-coding |
| Map location | 2p25.1 |
| Pascal p-value | 0.248 |
| Sherlock p-value | 0.57 |
| Fetal beta | 0.741 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_mGluR5 G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3547 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02466933 | 2 | 9770529 | YWHAQ | 1.85E-5 | -0.333 | 0.016 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs36781 | chr5 | 109289728 | YWHAQ | 10971 | 0.12 | trans |
Section II. Transcriptome annotation
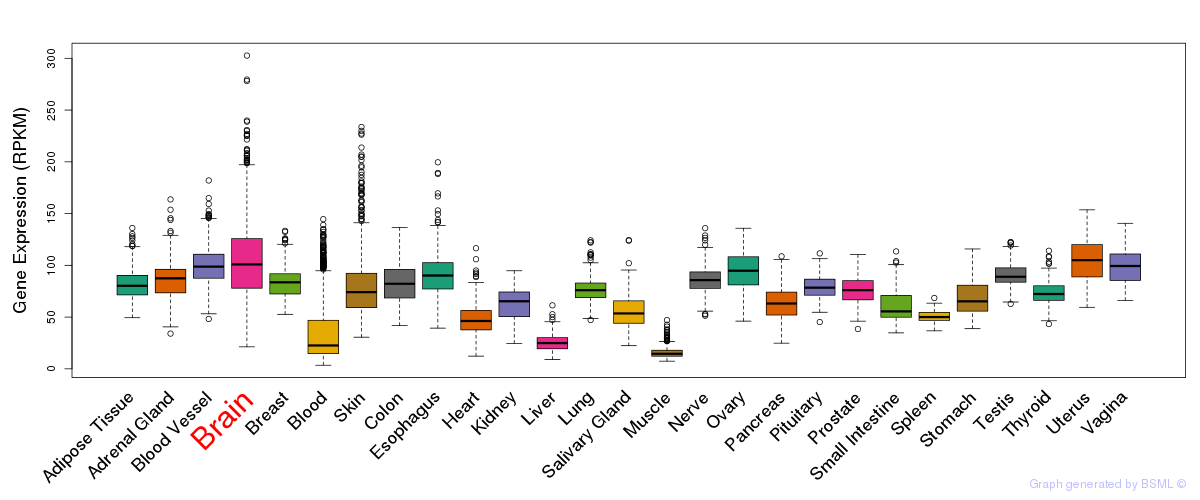
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIF2C | 0.97 | 0.69 |
| TACC3 | 0.96 | 0.73 |
| HJURP | 0.96 | 0.70 |
| PRC1 | 0.96 | 0.77 |
| RAD51 | 0.96 | 0.68 |
| BUB1B | 0.96 | 0.79 |
| GTSE1 | 0.96 | 0.73 |
| NCAPH | 0.96 | 0.73 |
| NUSAP1 | 0.96 | 0.71 |
| KIF20A | 0.96 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.40 | -0.66 |
| AF347015.33 | -0.40 | -0.67 |
| AF347015.31 | -0.40 | -0.64 |
| HLA-F | -0.40 | -0.54 |
| MT-CO2 | -0.40 | -0.65 |
| MT-CYB | -0.39 | -0.64 |
| TINAGL1 | -0.39 | -0.56 |
| C5orf53 | -0.39 | -0.48 |
| PTH1R | -0.39 | -0.51 |
| AF347015.8 | -0.38 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019904 | protein domain specific binding | IEA | - | |
| GO:0047485 | protein N-terminus binding | IPI | 11984006 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 15163635 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 18029348 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Reconstituted Complex | BioGRID | 11266503 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | Affinity Capture-Western | BioGRID | 14970201 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD,BioGRID | 9369453 |
| BAX | BCL2L4 | BCL2-associated X protein | - | HPRD,BioGRID | 12426317 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD | 8668348 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | Affinity Capture-MS | BioGRID | 17353931 |
| C22orf9 | - | chromosome 22 open reading frame 9 | Affinity Capture-MS | BioGRID | 17353931 |
| CABIN1 | CAIN | KIAA0330 | PPP3IN | calcineurin binding protein 1 | YWHAQ (14-3-3-theta) interacts with phosphorylated CABIN1. This interaction was modelled on a demonstrated interaction between mouse 14-3-3-theta and human CABIN1. | BIND | 15902271 |
| CALML5 | CLSP | calmodulin-like 5 | Affinity Capture-MS | BioGRID | 17353931 |
| CAPN3 | CANP3 | CANPL3 | LGMD2 | LGMD2A | MGC10767 | MGC11121 | MGC14344 | MGC4403 | nCL-1 | p94 | calpain 3, (p94) | - | HPRD | 15324660 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 8663231 |11024037 |
| CBWD2 | - | COBW domain containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CCAR1 | CARP-1 | CARP1 | MGC44628 | RP11-437A18.1 | cell division cycle and apoptosis regulator 1 | Affinity Capture-Western | BioGRID | 12816952 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 10713667 |
| CDC2L1 | CDC2L2 | CDK11 | CDK11-p110 | CDK11-p46 | CDK11-p58 | CLK-1 | FLJ59152 | PK58 | p58 | p58CDC2L1 | p58CLK-1 | cell division cycle 2-like 1 (PITSLRE proteins) | CDK11-p110 interacts with 14-3-3-theta. | BIND | 15883043 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD,BioGRID | 12042314 |
| CRTC2 | RP11-422P24.6 | TORC2 | CREB regulated transcription coactivator 2 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15454081 |17353931 |
| DAPK1 | DAPK | DKFZp781I035 | death-associated protein kinase 1 | Affinity Capture-Western | BioGRID | 12911633 |
| DDX6 | FLJ36338 | HLR2 | P54 | RCK | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | Affinity Capture-MS | BioGRID | 17353931 |
| DENND4A | FLJ33949 | IRLB | KIAA0476 | MYCPBP | DENN/MADD domain containing 4A | Affinity Capture-MS | BioGRID | 17353931 |
| EDC3 | FLJ21128 | FLJ31777 | LSM16 | YJDC | YJEFN2 | enhancer of mRNA decapping 3 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| EML3 | ELP95 | FLJ35827 | MGC111422 | echinoderm microtubule associated protein like 3 | Affinity Capture-MS | BioGRID | 17353931 |
| EPB41L2 | 4.1-G | DKFZp781D1972 | DKFZp781H1755 | erythrocyte membrane protein band 4.1-like 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EPB41L3 | 4.1B | DAL-1 | DAL1 | FLJ37633 | KIAA0987 | erythrocyte membrane protein band 4.1-like 3 | Affinity Capture-MS | BioGRID | 17353931 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Reconstituted Complex | BioGRID | 11266503 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | Reconstituted Complex | BioGRID | 11266503 |
| FAM83G | FLJ41564 | family with sequence similarity 83, member G | Affinity Capture-MS | BioGRID | 17353931 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD | 10066823 |
| FSHR | FSHRO | LGR1 | MGC141667 | MGC141668 | ODG1 | follicle stimulating hormone receptor | - | HPRD | 15196694 |
| FUSIP1 | FUSIP2 | NSSR | SFRS13 | SRp38 | SRrp40 | TASR | TASR1 | TASR2 | FUS interacting protein (serine/arginine-rich) 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FXYD1 | MGC44983 | PLM | FXYD domain containing ion transport regulator 1 | Reconstituted Complex | BioGRID | 14597563 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | Affinity Capture-Western Two-hybrid | BioGRID | 15367659 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 15324660 |
| IRS2 | - | insulin receptor substrate 2 | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA1409 | FLJ43337 | KIAA1409 | Affinity Capture-MS | BioGRID | 17353931 |
| KIF23 | CHO1 | KNSL5 | MKLP-1 | MKLP1 | kinesin family member 23 | Affinity Capture-MS | BioGRID | 17353931 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | Affinity Capture-MS | BioGRID | 17353931 |
| KIF5C | FLJ44735 | KIAA0531 | KINN | MGC111478 | NKHC | NKHC-2 | NKHC2 | kinesin family member 5C | Affinity Capture-MS | BioGRID | 17353931 |
| KLC2 | FLJ12387 | kinesin light chain 2 | Affinity Capture-MS | BioGRID | 17353931 |
| KLC3 | KLC2 | KLC2L | KLCt | KNS2B | kinesin light chain 3 | - | HPRD | 15324660 |
| KLC4 | KNSL8 | MGC111777 | bA387M24.3 | kinesin light chain 4 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT18 | CYK18 | K18 | keratin 18 | Affinity Capture-Western | BioGRID | 9524113 |
| LARP1 | KIAA0731 | LARP | MGC19556 | La ribonucleoprotein domain family, member 1 | Affinity Capture-MS | BioGRID | 17353931 |
| LUC7L2 | CGI-59 | CGI-74 | FLJ10657 | LUC7B2 | LUC7-like 2 (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | - | HPRD,BioGRID | 11984006 |
| MARK2 | EMK1 | MGC99619 | PAR-1 | Par1b | MAP/microtubule affinity-regulating kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| MARK3 | CTAK1 | KP78 | PAR1A | MAP/microtubule affinity-regulating kinase 3 | Affinity Capture-MS | BioGRID | 17353931 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Reconstituted Complex | BioGRID | 11266503 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11433030 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | Affinity Capture-MS | BioGRID | 17353931 |
| MPRIP | KIAA0864 | M-RIP | RHOIP3 | p116Rip | myosin phosphatase Rho interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | Reconstituted Complex | BioGRID | 11266503 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | Reconstituted Complex | BioGRID | 11266503 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 10611249 |
| NFATC4 | NF-ATc4 | NFAT3 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | - | HPRD,BioGRID | 10611249 |
| NIF3L1 | ALS2CR1 | CALS-7 | MDS015 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | - | HPRD,BioGRID | 15163635 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | Co-localization Phenotypic Suppression Reconstituted Complex | BioGRID | 11266503 |
| PAK4 | - | p21 protein (Cdc42/Rac)-activated kinase 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PAK6 | PAK5 | p21 protein (Cdc42/Rac)-activated kinase 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PANK2 | C20orf48 | FLJ17232 | HARP | HSS | MGC15053 | NBIA1 | PKAN | pantothenate kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD,BioGRID | 12177059 |
| PFKFB2 | DKFZp781D2217 | MGC138308 | MGC138310 | PFK-2/FBPase-2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | - | HPRD,BioGRID | 12853467 |
| PI4KB | PI4K-BETA | PI4KIIIbeta | PI4Kbeta | PIK4CB | pi4K92 | phosphatidylinositol 4-kinase, catalytic, beta | Affinity Capture-MS | BioGRID | 17353931 |
| PI4KB | PI4K-BETA | PI4KIIIbeta | PI4Kbeta | PIK4CB | pi4K92 | phosphatidylinositol 4-kinase, catalytic, beta | - | HPRD | 15324660 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Reconstituted Complex | BioGRID | 8663231 |
| PIK3CG | PI3CG | PI3K | PI3Kgamma | PIK3 | phosphoinositide-3-kinase, catalytic, gamma polypeptide | Affinity Capture-Western | BioGRID | 8663231 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Affinity Capture-MS | BioGRID | 17353931 |
| PPFIBP1 | L2 | hSGT2 | hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) | Affinity Capture-MS | BioGRID | 17353931 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 10620507 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10092600 |10831594 |
| RABEP1 | RAB5EP | RABPT5 | rabaptin, RAB GTPase binding effector protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-MS Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8663231 |10620507 |10757792 |17353931 |
| RAI14 | DKFZp564G013 | KIAA1334 | NORPEG | RAI13 | retinoic acid induced 14 | Affinity Capture-MS | BioGRID | 17353931 |
| RASGRP3 | GRP3 | KIAA0846 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | Affinity Capture-MS | BioGRID | 17353931 |
| REM1 | GD:REM | GES | MGC48669 | REM | RAS (RAD and GEM)-like GTP-binding 1 | - | HPRD,BioGRID | 10441394 |
| RPL21 | DKFZp686C06101 | FLJ27458 | L21 | MGC104274 | MGC104275 | MGC71252 | ribosomal protein L21 | Affinity Capture-MS | BioGRID | 17353931 |
| SAMD4B | FLJ10211 | MGC99832 | SMGB | sterile alpha motif domain containing 4B | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS3 | SRp20 | splicing factor, arginine/serine-rich 3 | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS6 | B52 | FLJ08061 | MGC5045 | SRP55 | splicing factor, arginine/serine-rich 6 | Affinity Capture-MS | BioGRID | 17353931 |
| SH3BP4 | BOG25 | TTP | SH3-domain binding protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| SPIRE1 | MGC150621 | MGC150622 | Spir-1 | spire homolog 1 (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| SUV420H1 | CGI-85 | CGI85 | KMT5B | MGC118906 | MGC118909 | MGC21161 | MGC703 | suppressor of variegation 4-20 homolog 1 (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | Reconstituted Complex Two-hybrid | BioGRID | 10835362 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Reconstituted Complex | BioGRID | 11266503 |
| TJP2 | MGC26306 | X104 | ZO-2 | ZO2 | tight junction protein 2 (zona occludens 2) | Affinity Capture-MS | BioGRID | 17353931 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Hamartin interacts with 14-3-3-tau. This interaction is modeled on demonstrated interaction between human hamartin and mouse 14-3-3-tau isoform. | BIND | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-MS | BioGRID | 17353931 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Tumor suppressor protein tuberin interacts with 14-3-3-tau isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-tau isoform. | BIND | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD | 12438239 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 15324660 |
| UCP2 | BMIQ4 | SLC25A8 | UCPH | uncoupling protein 2 (mitochondrial, proton carrier) | - | HPRD,BioGRID | 10785390 |
| UCP2 | BMIQ4 | SLC25A8 | UCPH | uncoupling protein 2 (mitochondrial, proton carrier) | UCP2 interacts with 14.3.3-theta. | BIND | 10785390 |
| UCP3 | SLC25A9 | uncoupling protein 3 (mitochondrial, proton carrier) | UCP3 interacts with 14.3.3-theta. | BIND | 10785390 |
| UCP3 | SLC25A9 | uncoupling protein 3 (mitochondrial, proton carrier) | - | HPRD,BioGRID | 10785390 |
| USP8 | FLJ34456 | HumORF8 | KIAA0055 | MGC129718 | UBPY | ubiquitin specific peptidase 8 | Affinity Capture-MS | BioGRID | 17353931 |
| WDR77 | HKMT1069 | MEP50 | MGC2722 | Nbla10071 | RP11-552M11.3 | p44 | WD repeat domain 77 | - | HPRD | 15324660 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 12535517 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| KOMMAGANI TP63 GAMMA TARGETS | 9 | 7 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN | 85 | 56 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 400 | 406 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-129-5p | 944 | 950 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-139 | 934 | 940 | m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| hsa-miR-139brain | UCUACAGUGCACGUGUCU | ||||
| miR-15/16/195/424/497 | 617 | 623 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 693 | 699 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-203.1 | 225 | 231 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-320 | 138 | 144 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-376c | 972 | 978 | 1A | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-378* | 811 | 818 | 1A,m8 | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-543 | 961 | 967 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.