| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP
| 285 | 181 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN
| 526 | 357 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP
| 51 | 36 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN
| 182 | 111 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN
| 244 | 147 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN
| 320 | 184 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN
| 205 | 127 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN
| 43 | 29 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP
| 98 | 58 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS B DN
| 8 | 5 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS
| 355 | 243 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN
| 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS
| 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3
| 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP
| 64 | 40 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS
| 183 | 115 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP
| 60 | 42 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN
| 246 | 180 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP
| 268 | 157 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP
| 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3
| 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4
| 94 | 69 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4
| 100 | 62 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN
| 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN
| 281 | 179 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR DN
| 48 | 31 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP
| 41 | 29 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP
| 260 | 174 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP
| 147 | 98 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP
| 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP
| 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN
| 245 | 150 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED
| 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN
| 113 | 76 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP
| 101 | 65 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN
| 253 | 192 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM
| 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING
| 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT
| 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA
| 208 | 107 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP
| 297 | 194 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
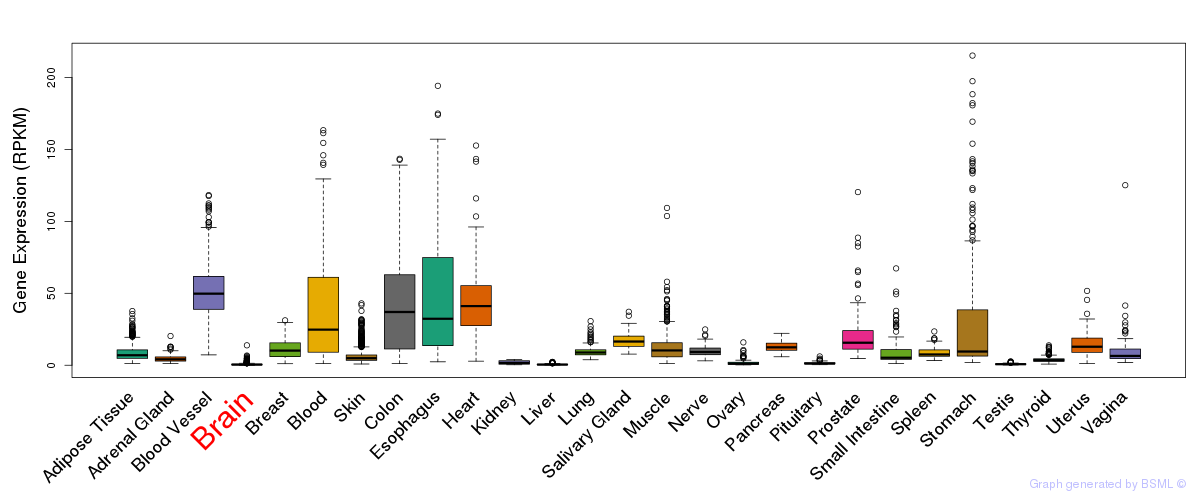
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation