Gene Page: PARP1
Summary ?
| GeneID | 142 |
| Symbol | PARP1 |
| Synonyms | ADPRT|ADPRT 1|ADPRT1|ARTD1|PARP|PARP-1|PPOL|pADPRT-1 |
| Description | poly(ADP-ribose) polymerase 1 |
| Reference | MIM:173870|HGNC:HGNC:270|Ensembl:ENSG00000143799|HPRD:01435|Vega:OTTHUMG00000037556 |
| Gene type | protein-coding |
| Map location | 1q41-q42 |
| Sherlock p-value | 0.664 |
| Fetal beta | -0.497 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0455 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17127702 | 1 | 226594323 | PARP1 | 5.818E-4 | 0.444 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
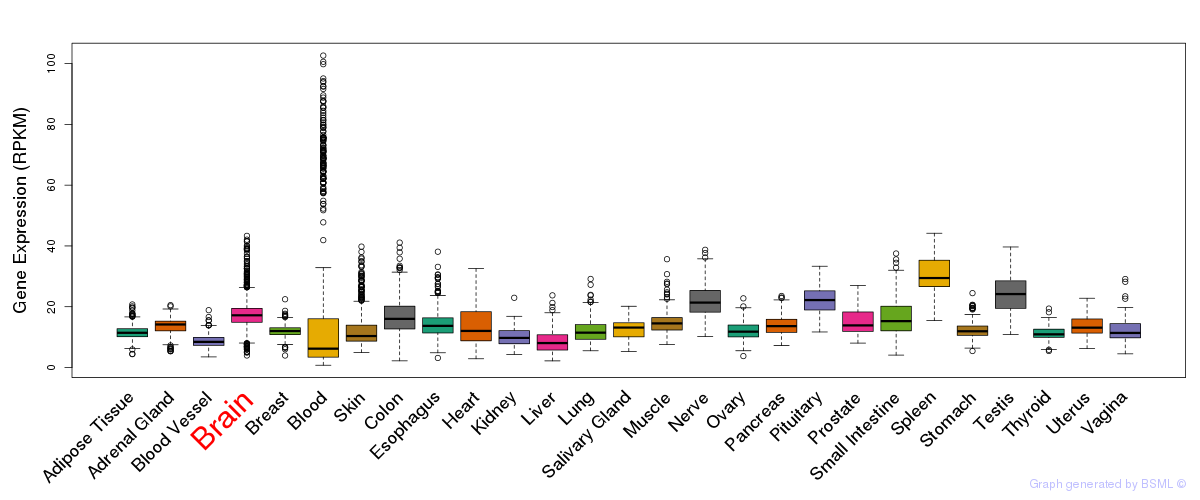
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003950 | NAD+ ADP-ribosyltransferase activity | IDA | 16107709 | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003677 | DNA binding | TAS | 2513174 | |
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 14734561 | |
| GO:0051287 | NAD binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0047485 | protein N-terminus binding | IPI | 16107709 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006471 | protein amino acid ADP-ribosylation | TAS | 2513174 | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 10944198 | |
| GO:0006281 | DNA repair | TAS | 10958667 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 2513174 | |
| GO:0005635 | nuclear envelope | IDA | 9518481 | |
| GO:0005730 | nucleolus | IDA | 9518481 |16107709 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APTX | AOA | AOA1 | AXA1 | EAOH | EOAHA | FHA-HIT | FLJ20157 | MGC1072 | aprataxin | - | HPRD,BioGRID | 15044383 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 11790116 |
| BUB3 | BUB3L | hBUB3 | budding uninhibited by benzimidazoles 3 homolog (yeast) | - | HPRD,BioGRID | 12011073 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | PARP interacts with Caspase-3. This interaction was modeled on a demonstrated interaction between human PARP and caspase-3 from an unspecified source. | BIND | 15186778 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | PARP interacts with caspase-7. This interaction was modelled on a demonstrated interaction between human PARP and fly drICE. | BIND | 15580265 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | - | HPRD,BioGRID | 12930846 |
| CENPA | CENP-A | centromere protein A | - | HPRD,BioGRID | 12011073 |
| CENPB | - | centromere protein B, 80kDa | - | HPRD,BioGRID | 12011073 |
| MED6 | NY-REN-28 | mediator complex subunit 6 | PARP-1 interacts with Med6. | BIND | 15808511 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | - | HPRD,BioGRID | 10744766 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-MS | BioGRID | 12519782 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 11590148 |
| PARP2 | ADPRT2 | ADPRTL2 | ADPRTL3 | PARP-2 | pADPRT-2 | poly (ADP-ribose) polymerase 2 | - | HPRD,BioGRID | 12217960 |
| PARP3 | ADPRT3 | ADPRTL2 | ADPRTL3 | IRT1 | hPARP-3 | pADPRT-3 | poly (ADP-ribose) polymerase family, member 3 | - | HPRD,BioGRID | 12640039 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | Affinity Capture-Western | BioGRID | 12930846 |
| POLA1 | DKFZp686K1672 | POLA | p180 | polymerase (DNA directed), alpha 1, catalytic subunit | - | HPRD,BioGRID | 9518481 |9563011 |
| POLA2 | FLJ21662 | FLJ37250 | polymerase (DNA directed), alpha 2 (70kD subunit) | - | HPRD,BioGRID | 9518481 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD | 9537509 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD | 9398855 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | PARP-1 interacts with RAR-alpha. | BIND | 15808511 |
| RBM14 | COAA | DKFZp779J0927 | MGC15912 | MGC31756 | PSP2 | SIP | SYTIP1 | TMEM137 | RNA binding motif protein 14 | - | HPRD,BioGRID | 11443112 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 11590148 |
| RPS3A | FTE1 | MFTL | MGC23240 | ribosomal protein S3A | - | HPRD,BioGRID | 11790116 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD | 10082530 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| SWAP70 | FLJ39540 | HSPC321 | KIAA0640 | SWAP-70 | SWAP-70 protein | - | HPRD,BioGRID | 9642267 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Affinity Capture-MS | BioGRID | 16407974 |
| TERF2IP | DRIP5 | RAP1 | telomeric repeat binding factor 2, interacting protein | Affinity Capture-MS | BioGRID | 15100233 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | PARP-1 interacts with TR-alpha. This interaction was modeled on a demonstrated interaction between human PARP-1 and TR-alpha from an unspecified species. | BIND | 15808511 |
| TOP1 | TOPI | topoisomerase (DNA) I | PARP-1 interacts with Topo I | BIND | 15247263 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9565608 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | WRN interacts with PARP1. | BIND | 14734561 |
| XRCC1 | RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9584196 |15044383 |
| XRCC1 | RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 | - | HPRD | 9584196 |14500814 |
| ZNF423 | Ebfaz | KIAA0760 | MGC138520 | MGC138522 | OAZ | Roaz | ZFP423 | Zfp104 | zinc finger protein 423 | - | HPRD,BioGRID | 14623329 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASE EXCISION REPAIR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA D4GDI PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| SA CASPASE CASCADE | 19 | 13 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH PROLIFERATION | 147 | 80 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC REGULATORS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS UP | 79 | 46 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| LY AGING PREMATURE DN | 30 | 17 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| SYED ESTRADIOL RESPONSE | 19 | 15 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ABDELMOHSEN ELAVL4 TARGETS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 326 | 332 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 326 | 332 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.