Gene Page: CYP1B1
Summary ?
| GeneID | 1545 |
| Symbol | CYP1B1 |
| Synonyms | CP1B|CYPIB1|GLC3A|P4501B1 |
| Description | cytochrome P450 family 1 subfamily B member 1 |
| Reference | MIM:601771|HGNC:HGNC:2597|Ensembl:ENSG00000138061|HPRD:03464|Vega:OTTHUMG00000100970 |
| Gene type | protein-coding |
| Map location | 2p22.2 |
| Pascal p-value | 0.54 |
| Sherlock p-value | 0.01 |
| Fetal beta | -0.552 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs848206 | chr1 | 16252995 | CYP1B1 | 1545 | 0.02 | trans | ||
| rs848215 | chr1 | 16267146 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs1320385 | chr1 | 54064468 | CYP1B1 | 1545 | 0.04 | trans | ||
| rs12036675 | chr1 | 54066317 | CYP1B1 | 1545 | 0.04 | trans | ||
| rs12062528 | chr1 | 54070830 | CYP1B1 | 1545 | 0.06 | trans | ||
| rs4522651 | chr2 | 34048 | CYP1B1 | 1545 | 1.931E-4 | trans | ||
| rs2839839 | chr2 | 51691483 | CYP1B1 | 1545 | 0.03 | trans | ||
| rs6739534 | chr2 | 51702264 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs1911135 | chr2 | 228688488 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs2922180 | chr3 | 125280472 | CYP1B1 | 1545 | 5.091E-6 | trans | ||
| rs2971271 | chr3 | 125450402 | CYP1B1 | 1545 | 0.05 | trans | ||
| rs2976809 | chr3 | 125451738 | CYP1B1 | 1545 | 0.05 | trans | ||
| rs1395157 | chr5 | 40055537 | CYP1B1 | 1545 | 0.05 | trans | ||
| rs16888672 | chr5 | 58126580 | CYP1B1 | 1545 | 0.16 | trans | ||
| rs16894149 | chr5 | 61013724 | CYP1B1 | 1545 | 0.06 | trans | ||
| rs17558509 | chr5 | 125107290 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs6595634 | chr5 | 125113615 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs185323 | chr5 | 154212519 | CYP1B1 | 1545 | 9.615E-4 | trans | ||
| rs702743 | chr5 | 154271390 | CYP1B1 | 1545 | 0.04 | trans | ||
| rs816736 | chr5 | 154271947 | CYP1B1 | 1545 | 1.061E-7 | trans | ||
| rs816748 | chr5 | 154274691 | CYP1B1 | 1545 | 9.615E-4 | trans | ||
| rs863650 | chr5 | 154281215 | CYP1B1 | 1545 | 0.04 | trans | ||
| rs348737 | chr5 | 154297807 | CYP1B1 | 1545 | 0.19 | trans | ||
| rs1390602 | chr5 | 154385024 | CYP1B1 | 1545 | 9.615E-4 | trans | ||
| rs17570361 | chr5 | 154397675 | CYP1B1 | 1545 | 9.615E-4 | trans | ||
| rs9389245 | chr6 | 135265648 | CYP1B1 | 1545 | 0.04 | trans | ||
| rs2529439 | chr7 | 30643747 | CYP1B1 | 1545 | 0.05 | trans | ||
| rs2240404 | chr7 | 30707301 | CYP1B1 | 1545 | 0.17 | trans | ||
| rs4723002 | chr7 | 30725700 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs4723003 | chr7 | 30725740 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs7841407 | chr8 | 9243427 | CYP1B1 | 1545 | 0 | trans | ||
| rs2739708 | chr8 | 17992280 | CYP1B1 | 1545 | 0.16 | trans | ||
| rs714131 | chr8 | 17992832 | CYP1B1 | 1545 | 0.17 | trans | ||
| rs16876827 | chr8 | 30148763 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs10955813 | chr8 | 118415080 | CYP1B1 | 1545 | 0.13 | trans | ||
| rs10969120 | chr9 | 29400711 | CYP1B1 | 1545 | 0.13 | trans | ||
| rs4743989 | chr9 | 97859191 | CYP1B1 | 1545 | 0.02 | trans | ||
| rs356660 | chr9 | 98006134 | CYP1B1 | 1545 | 0.16 | trans | ||
| rs10886877 | chr10 | 85425690 | CYP1B1 | 1545 | 0.1 | trans | ||
| rs7071168 | chr10 | 95829674 | CYP1B1 | 1545 | 0.05 | trans | ||
| rs4612791 | chr11 | 40768423 | CYP1B1 | 1545 | 0.07 | trans | ||
| rs11115813 | chr12 | 83808239 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs17054673 | chr13 | 58610114 | CYP1B1 | 1545 | 0.19 | trans | ||
| snp_a-2256421 | 0 | CYP1B1 | 1545 | 0.13 | trans | |||
| rs17070066 | chr13 | 58661554 | CYP1B1 | 1545 | 0.17 | trans | ||
| rs6587203 | chr17 | 19671457 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs205113 | chr17 | 19673879 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs205111 | chr17 | 19675103 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs205110 | chr17 | 19675587 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs205093 | chr17 | 19685712 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs157406 | chr17 | 19710650 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs157402 | chr17 | 19720588 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs157396 | chr17 | 19726832 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs157397 | chr17 | 19729494 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs8074258 | 0 | CYP1B1 | 1545 | 0.09 | trans | |||
| rs172186 | chr17 | 19780658 | CYP1B1 | 1545 | 0.01 | trans | ||
| rs281334 | chr17 | 19782079 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs4586510 | chr17 | 19783973 | CYP1B1 | 1545 | 0.09 | trans | ||
| rs9959804 | chr18 | 20264142 | CYP1B1 | 1545 | 0.01 | trans |
Section II. Transcriptome annotation
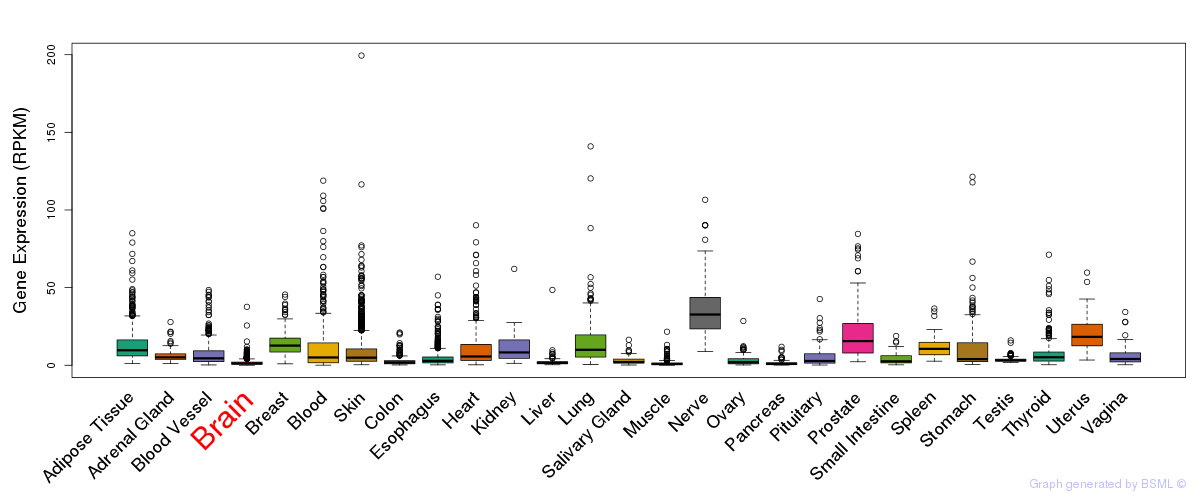
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STEROID HORMONE BIOSYNTHESIS | 55 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG METABOLISM OF XENOBIOTICS BY CYTOCHROME P450 | 70 | 44 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOGENOUS STEROLS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN GRANULOCYTE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION GRANULOCYTE DN | 17 | 13 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL DIVIDING UP | 13 | 7 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| LI CISPLATIN RESISTANCE UP | 28 | 20 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| WOTTON RUNX TARGETS UP | 21 | 14 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER F | 54 | 32 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| JI METASTASIS REPRESSED BY STK11 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA MAST CELL | 46 | 34 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| CHUANG OXIDATIVE STRESS RESPONSE UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 12 | 36 | 18 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS DN | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BUCKANOVICH T LYMPHOCYTE HOMING ON TUMOR UP | 24 | 11 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| VISALA AGING LYMPHOCYTE UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |