Gene Page: EP300
Summary ?
| GeneID | 2033 |
| Symbol | EP300 |
| Synonyms | KAT3B|RSTS2|p300 |
| Description | E1A binding protein p300 |
| Reference | MIM:602700|HGNC:HGNC:3373|Ensembl:ENSG00000100393|HPRD:04078|Vega:OTTHUMG00000150937 |
| Gene type | protein-coding |
| Map location | 22q13.2 |
| Pascal p-value | 6.924E-6 |
| Sherlock p-value | 0.807 |
| Fetal beta | 0.728 |
| eGene | Cerebellar Hemisphere Myers' cis & trans Meta |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0862 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EP300 | chr22 | 41546041 | C | G | NM_001429 | p.886P>A | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | EP300 | 2033 | 1.229E-4 | trans | ||
| rs9810143 | chr3 | 5060209 | EP300 | 2033 | 0.11 | trans | ||
| rs11186601 | chr10 | 93287199 | EP300 | 2033 | 0.11 | trans | ||
| rs16955618 | chr15 | 29937543 | EP300 | 2033 | 0 | trans | ||
| rs16990575 | chrX | 32691327 | EP300 | 2033 | 0.18 | trans |
Section II. Transcriptome annotation
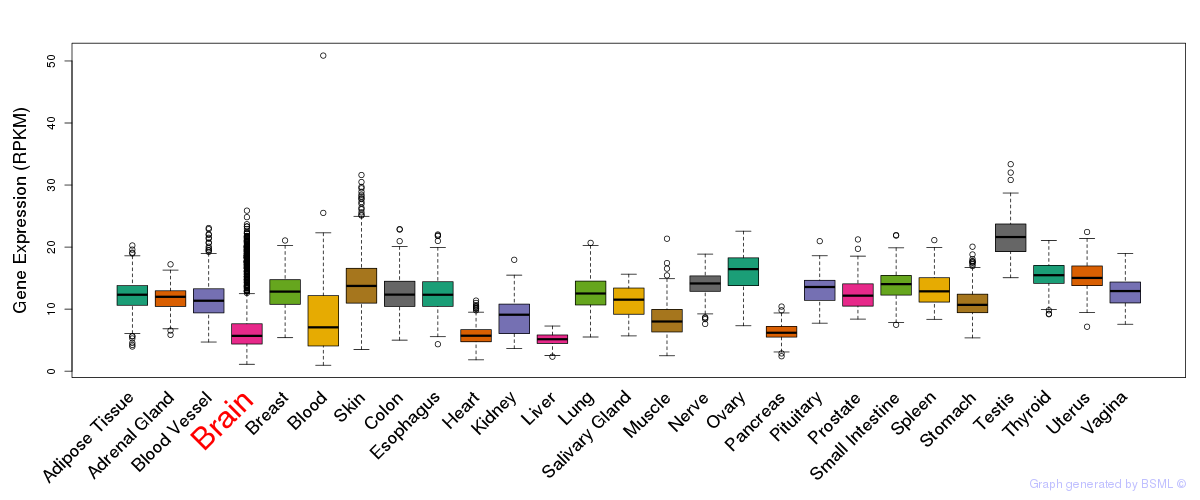
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DLG5 | 0.96 | 0.95 |
| EHMT1 | 0.95 | 0.90 |
| ZXDC | 0.95 | 0.94 |
| NUP62 | 0.95 | 0.97 |
| PFAS | 0.95 | 0.97 |
| RBM15B | 0.95 | 0.97 |
| CD276 | 0.95 | 0.93 |
| DNMT1 | 0.95 | 0.97 |
| GPR161 | 0.95 | 0.94 |
| PHF2 | 0.94 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.73 | -0.80 |
| AIFM3 | -0.71 | -0.79 |
| HLA-F | -0.71 | -0.78 |
| AF347015.31 | -0.70 | -0.92 |
| AF347015.27 | -0.70 | -0.92 |
| S100B | -0.70 | -0.87 |
| ALDOC | -0.70 | -0.73 |
| MT-CO2 | -0.69 | -0.93 |
| AF347015.33 | -0.69 | -0.90 |
| TSC22D4 | -0.68 | -0.80 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 10893273 | |
| GO:0003713 | transcription coactivator activity | IDA | 12435739 |12586840 | |
| GO:0004402 | histone acetyltransferase activity | IDA | 12040021 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008022 | protein C-terminus binding | TAS | 10205054 | |
| GO:0016407 | acetyltransferase activity | IMP | 12435739 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | IEA | neuron (GO term level: 8) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 10205054 |
| GO:0001756 | somitogenesis | IEA | - | |
| GO:0001666 | response to hypoxia | IDA | 15261140 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007165 | signal transduction | NAS | 15261140 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0006915 | apoptosis | IMP | 9194565 | |
| GO:0018076 | N-terminal peptidyl-lysine acetylation | IDA | 12435739 | |
| GO:0051091 | positive regulation of transcription factor activity | IDA | 10518217 | |
| GO:0016573 | histone acetylation | IEA | - | |
| GO:0042592 | homeostatic process | TAS | 15261140 | |
| GO:0030324 | lung development | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000123 | histone acetyltransferase complex | IEA | - | |
| GO:0005634 | nucleus | IDA | 9194565 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALX1 | CART1 | ALX homeobox 1 | - | HPRD | 12929931 |
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | Reconstituted Complex | BioGRID | 14645221 |
| ASCL1 | ASH1 | HASH1 | MASH1 | bHLHa46 | achaete-scute complex homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 11564735 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | Reconstituted Complex Two-hybrid | BioGRID | 10497212 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | The cIAP-2 promoter interacts with p300. | BIND | 15494311 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 10655477 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | p300 interacts with BRCA1. | BIND | 10655477 |
| CARM1 | PRMT4 | coactivator-associated arginine methyltransferase 1 | CARM1 interacts with p300. | BIND | 15616592 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 11788592 |
| CDX2 | CDX-3 | CDX3 | caudal type homeobox 2 | - | HPRD,BioGRID | 10506141 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 9343424 |
| CITED1 | MSG1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | - | HPRD,BioGRID | 10722728 |
| CITED2 | MRG1 | P35SRJ | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9887100 |10593900 |11744733 |12586840 |
| CITED2 | MRG1 | P35SRJ | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | - | HPRD | 9887100 |12586840 |
| CITED4 | - | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | - | HPRD,BioGRID | 11744733 |
| CLOCK | KAT13D | KIAA0334 | bHLHe8 | clock homolog (mouse) | Reconstituted Complex | BioGRID | 14645221 |
| CNTN2 | AXT | DKFZp781D102 | FLJ42746 | MGC157722 | TAG-1 | TAX | TAX1 | contactin 2 (axonal) | Phenotypic Suppression | BioGRID | 11264182 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Affinity Capture-Western | BioGRID | 10506141 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | CTBP1 interacts with EP300 (p300) | BIND | 15834423 |
| CXCL10 | C7 | IFI10 | INP10 | IP-10 | SCYB10 | crg-2 | gIP-10 | mob-1 | chemokine (C-X-C motif) ligand 10 | p300 interacts with promoter IP-10. | BIND | 15315758 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | Affinity Capture-Western | BioGRID | 12527917 |
| DTX1 | hDx-1 | deltex homolog 1 (Drosophila) | - | HPRD,BioGRID | 11564735 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | p300 interacts with Egr1 promoter. | BIND | 15225550 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | - | HPRD,BioGRID | 9806899 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | p300 interacts with Egr1. | BIND | 15225550 |
| EID1 | C15orf3 | CRI1 | EID-1 | IRO45620 | MGC138883 | MGC138884 | PNAS-22 | PTD014 | RBP21 | EP300 interacting inhibitor of differentiation 1 | - | HPRD,BioGRID | 11073989 |11073990 |
| EID2 | CRI2 | EID-2 | MGC20452 | EP300 interacting inhibitor of differentiation 2 | - | HPRD | 14585496 |
| ELK1 | - | ELK1, member of ETS oncogene family | Affinity Capture-Western Phenotypic Enhancement Reconstituted Complex | BioGRID | 12514134 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Biochemical Activity | BioGRID | 10567538 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | EP300 (p300) autoacetylates. This interaction was modeled on a demonstrated interaction using EP300 from an unspecified species. | BIND | 14761960 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11782371 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | EP300 (p300) acetylates ESR1 (ER-alpha). This interaction was modeled on a demonstrated interaction between EP300 from an unspecified species and ESR1 from an unspecified species. | BIND | 14761960 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD,BioGRID | 10942770 |
| ETS2 | ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | - | HPRD,BioGRID | 10942770 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | - | HPRD,BioGRID | 11430825 |
| GATA6 | - | GATA binding protein 6 | - | HPRD,BioGRID | 10851229 |
| GPS2 | AMF-1 | MGC104294 | MGC119287 | MGC119288 | MGC119289 | G protein pathway suppressor 2 | - | HPRD,BioGRID | 10846067 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | p300 interacts with TFIIB. This interaction is modelled on a demonstrated interaction between human p300 and TFIIB from an unspecified species. | BIND | 8621548 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | EP300 (p300) interacts with H3F3A (histone 3). | BIND | 15834423 |
| HAND2 | DHAND2 | FLJ16260 | Hed | MGC125303 | MGC125304 | Thing2 | bHLHa26 | dHand | heart and neural crest derivatives expressed 2 | - | HPRD,BioGRID | 11994297 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 11823643 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | EP300 (p300) interacts with HIST4H4 (histone 4) | BIND | 15834423 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 11978637 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | - | HPRD,BioGRID | 11909954 |
| IL8 | CXCL8 | GCP-1 | GCP1 | LECT | LUCT | LYNAP | MDNCF | MONAP | NAF | NAP-1 | NAP1 | interleukin 8 | The IL-8 promoter interacts with p300. | BIND | 15494311 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | - | HPRD,BioGRID | 12015309 |
| ING4 | MGC12557 | my036 | p29ING4 | inhibitor of growth family, member 4 | p300 interacts with p29ING4. | BIND | 12750254 |
| ING4 | MGC12557 | my036 | p29ING4 | inhibitor of growth family, member 4 | - | HPRD,BioGRID | 12750254 |
| ING5 | FLJ23842 | p28ING5 | inhibitor of growth family, member 5 | - | HPRD,BioGRID | 12750254 |
| ING5 | FLJ23842 | p28ING5 | inhibitor of growth family, member 5 | p300 interacts with p28ING5. | BIND | 12750254 |
| IRF2 | DKFZp686F0244 | IRF-2 | interferon regulatory factor 2 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 11304541 |
| JDP2 | JUNDM2 | Jun dimerization protein 2 | - | HPRD | 12101239 |
| JMY | FLJ37870 | MGC163496 | junction-mediating and regulatory protein | - | HPRD,BioGRID | 10518217 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Biochemical Activity | BioGRID | 12419806 |
| KLF5 | BTEB2 | CKLF | IKLF | Kruppel-like factor 5 (intestinal) | Affinity Capture-Western | BioGRID | 14612398 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD,BioGRID | 12446687 |
| MAF | MGC71685 | c-MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | p300 interacts with c-Maf. This interaction was modeled on a demonstrated interaction between c-Maf from an unspecified source and human p300. | BIND | 11943779 |
| MAF | MGC71685 | c-MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | - | HPRD,BioGRID | 11943779 |
| MAML1 | KIAA0200 | Mam-1 | Mam1 | mastermind-like 1 (Drosophila) | - | HPRD,BioGRID | 12050117 |12391150 |
| MAML1 | KIAA0200 | Mam-1 | Mam1 | mastermind-like 1 (Drosophila) | p300 interacts with MAML1. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human MAML1. | BIND | 12391150 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 9809062 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 12483531 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD,BioGRID | 12371907 |
| MEF2C | - | myocyte enhancer factor 2C | Phenotypic Enhancement Reconstituted Complex | BioGRID | 9001254 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | Affinity Capture-Western | BioGRID | 10825153 |10933397 |
| MGMT | - | O-6-methylguanine-DNA methyltransferase | - | HPRD,BioGRID | 11564893 |
| MPG | AAG | APNG | CRA36.1 | MDG | Mid1 | PIG11 | PIG16 | anpg | N-methylpurine-DNA glycosylase | EP300 (p300) acetylates MPG. This interaction was modeled on a demonstrated interaction between EP300 from an unspecified species and MPG from an unspecified species. | BIND | 14761960 |
| MYB | Cmyb | c-myb | c-myb_CDS | efg | v-myb myeloblastosis viral oncogene homolog (avian) | c-Myb interacts with p300. | BIND | 15691758 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | - | HPRD,BioGRID | 11733503 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 9001254 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | p300 interacts with MyoD. This interaction is modelled on a demonstrated interaction between human and mouse p300 and MyoD from mouse and an unspecified species. | BIND | 8621548 |
| N4BP2 | B3BP | FLJ10680 | KIAA1413 | NEDD4 binding protein 2 | - | HPRD,BioGRID | 12730195 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 11113179 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | NCOA1 (SRC-1) interacts with EP300 (p300). | BIND | 15688032 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | GRIP1 interacts with p300. This interaction was modeled on a demonstrated interaction between mouse GRIP1 and human p300. | BIND | 9920895 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | Affinity Capture-Western | BioGRID | 10655477 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | - | HPRD,BioGRID | 10823961 |
| NEIL2 | FLJ31644 | MGC2832 | MGC4505 | NEH2 | NEI2 | nei like 2 (E. coli) | - | HPRD | 15175427 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | - | HPRD,BioGRID | 10545951 |
| NEUROG1 | AKA | Math4C | NEUROD3 | bHLHa6 | ngn1 | neurogenin 1 | Phenotypic Enhancement | BioGRID | 11239394 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 9625762 |
| NPAS2 | FLJ23138 | MGC71151 | MOP4 | PASD4 | bHLHe9 | neuronal PAS domain protein 2 | Reconstituted Complex | BioGRID | 14645221 |
| NR4A1 | GFRP1 | HMR | MGC9485 | N10 | NAK-1 | NGFIB | NP10 | NUR77 | TR3 | nuclear receptor subfamily 4, group A, member 1 | - | HPRD,BioGRID | 12082103 |
| NUP98 | ADIR2 | NUP196 | NUP96 | nucleoporin 98kDa | Affinity Capture-Western | BioGRID | 9858599 |
| NUPR1 | COM1 | P8 | nuclear protein 1 | p8 interacts with and is acetylated by p300. | BIND | 11940591 |
| P53AIP1 | - | p53-regulated apoptosis-inducing protein 1 | p300 interacts with p53AIP1 promoter. | BIND | 15893728 |
| PAX6 | AN | AN2 | D11S812E | MGC17209 | MGDA | WAGR | paired box 6 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10506141 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 11268218 |
| PELP1 | HMX3 | MNAR | P160 | proline, glutamate and leucine rich protein 1 | - | HPRD,BioGRID | 11481323 |
| PIAS3 | FLJ14651 | ZMIZ5 | protein inhibitor of activated STAT, 3 | Affinity Capture-Western | BioGRID | 14691252 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western Co-purification | BioGRID | 9443979 |9710619 |
| POU3F2 | BRN2 | OCT7 | OTF7 | POUF3 | POU class 3 homeobox 2 | Brn-2 interacts with p300. | BIND | 11029584 |
| PPARA | MGC2237 | MGC2452 | NR1C1 | PPAR | hPPAR | peroxisome proliferator-activated receptor alpha | - | HPRD,BioGRID | 9407140 |
| PPARD | FAAR | MGC3931 | NR1C2 | NUC1 | NUCI | NUCII | PPAR-beta | PPARB | peroxisome proliferator-activated receptor delta | - | HPRD | 10567538 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | Reconstituted Complex Two-hybrid | BioGRID | 10944516 |12479814 |
| PROX1 | - | prospero homeobox 1 | - | HPRD,BioGRID | 11943779 |
| PROX1 | - | prospero homeobox 1 | p300 interacts with Prox-1. This interaction was modeled on a demonstrated interaction between Prox-1 from an unspecified source and human p300. | BIND | 11943779 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | - | HPRD,BioGRID | 11967287 |
| RAD23A | HHR23A | MGC111083 | RAD23 homolog A (S. cerevisiae) | hHR23A interacts with p300. This interaction was modelled on a demonstrated interaction between hHR23A from human and p300 from an unspecified species. | BIND | 11196199 |
| RBM14 | COAA | DKFZp779J0927 | MGC15912 | MGC31756 | PSP2 | SIP | SYTIP1 | TMEM137 | RNA binding motif protein 14 | - | HPRD,BioGRID | 11443112 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | p300 interacts with p65. | BIND | 9096323 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | p300 interacts with p65. | BIND | 15616592 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 9096323 |
| RORA | DKFZp686M2414 | MGC119326 | MGC119329 | NR1F1 | ROR1 | ROR2 | ROR3 | RZR-ALPHA | RZRA | RAR-related orphan receptor A | - | HPRD,BioGRID | 9862959 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | - | HPRD | 15138260 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD,BioGRID | 10673036 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | p300 interacts with Smad2. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human Smad2. | BIND | 9679056 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Affinity Capture-Western | BioGRID | 10199400 |11371641 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 11264182 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Smad3 interacts with p300. This interaction was modeled on a demonstrated interaction between Smad3 and p300 from an unspecified sources. | BIND | 10497242 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | SMAD3 interacts with EP300 (p300). | BIND | 15688032 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Affinity Capture-Western | BioGRID | 10887155 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | p300 interacts with Smad4 fragment. p300 interacting with full-length Smad4 could not be demonstrated in vitro. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human Smad4 fragment. | BIND | 9679056 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD,BioGRID | 12408818 |
| SNIP1 | FLJ12553 | RP3-423B22.3 | dJ423B22.2 | Smad nuclear interacting protein 1 | - | HPRD,BioGRID | 10887155 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10973497 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | - | HPRD,BioGRID | 8918891 |
| SS18 | MGC116875 | SSXT | SYT | SYT-SSX1 | SYT-SSX2 | synovial sarcoma translocation, chromosome 18 | - | HPRD,BioGRID | 11030627 |
| SS18L1 | CREST | KIAA0693 | LP2261 | MGC26711 | MGC78386 | synovial sarcoma translocation gene on chromosome 18-like 1 | - | HPRD,BioGRID | 14716005 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Affinity Capture-Western | BioGRID | 10464260 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Stat1-beta interacts with p300. This interaction is modelled on a demonstrated interaction between human Stat1-beta and p300 from an unspecified species. | BIND | 8986769 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | Reconstituted Complex | BioGRID | 10464260 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | Stat2 interacts with p300. | BIND | 10464260 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Stat3 interacts with p300. | BIND | 15653507 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 10205054 |
| STAT6 | D12S1644 | IL-4-STAT | STAT6B | STAT6C | signal transducer and activator of transcription 6, interleukin-4 induced | - | HPRD,BioGRID | 10454341 |
| SUB1 | MGC102747 | P15 | PC4 | p14 | SUB1 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11279157 |
| TACC2 | AZU-1 | ECTACC | transforming, acidic coiled-coil containing protein 2 | TACC2 interacts with p300. | BIND | 14767476 |
| TADA3L | ADA3 | FLJ20221 | FLJ21329 | hADA3 | transcriptional adaptor 3 (NGG1 homolog, yeast)-like | - | HPRD,BioGRID | 11707411 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490830 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | p300 interacts with TBP. | BIND | 8621548 |
| TCF12 | HEB | HTF4 | HsT17266 | bHLHb20 | transcription factor 12 | HEBb interacts with p300. | BIND | 15333839 |
| TCF12 | HEB | HTF4 | HsT17266 | bHLHb20 | transcription factor 12 | HEBa interacts with p300. This interaction was modeled on a demonstrated interaction between HEBb and p300. | BIND | 15333839 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD,BioGRID | 12435739 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 11864601 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | - | HPRD,BioGRID | 12586840 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | p300 interacts with pS2. | BIND | 10490106 |
| TGS1 | DKFZp762A163 | FLJ22995 | NCOA6IP | PIMT | PIPMT | trimethylguanosine synthase homolog (S. cerevisiae) | - | HPRD,BioGRID | 11912212 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 12371907 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p300 interacts with p53 | BIND | 15558054 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p300 interacts with p53. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human p53. | BIND | 15186775 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p300 interacts with and acetylates p53. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human p53. | BIND | 15933712 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 10942770 |
| TP73 | P73 | tumor protein p73 | p73-alpha interacts with p300.This interaction was modeled on a demonstrated interaction between human p73-alpha and p300 from an unspecified species. | BIND | 15175157 |
| TRERF1 | BCAR2 | FLJ21198 | HSA277276 | RAPA | RP1-139D8.5 | TReP-132 | dJ139D8.5 | transcriptional regulating factor 1 | - | HPRD,BioGRID | 11349124 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD,BioGRID | 10454579 |
| TSG101 | TSG10 | VPS23 | tumor susceptibility gene 101 | - | HPRD,BioGRID | 10440698 |
| TWIST1 | ACS3 | BPES2 | BPES3 | SCS | TWIST | bHLHa38 | twist homolog 1 (Drosophila) | - | HPRD,BioGRID | 10025406 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD | 8824223 |
| USF2 | FIP | bHLHb12 | upstream transcription factor 2, c-fos interacting | - | HPRD,BioGRID | 10434034 |
| XPA | XP1 | XPAC | xeroderma pigmentosum, complementation group A | Affinity Capture-Western | BioGRID | 11268218 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 7758944 |11486036 |
| ZBTB17 | MIZ-1 | ZNF151 | ZNF60 | pHZ-67 | zinc finger and BTB domain containing 17 | Affinity Capture-Western | BioGRID | 11283613 |
| ZRANB2 | DKFZp686J1831 | DKFZp686N09117 | FLJ41119 | ZIS | ZIS1 | ZIS2 | ZNF265 | zinc finger, RAN-binding domain containing 2 | - | HPRD | 11448987 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RELA PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VDR PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL7 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM1 PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 22 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 989 | 995 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-132/212 | 1065 | 1072 | 1A,m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-142-5p | 417 | 423 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-148/152 | 259 | 265 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 416 | 422 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 733 | 739 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-186 | 559 | 565 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-200bc/429 | 669 | 675 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 436 | 442 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-217 | 128 | 134 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-22 | 727 | 733 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-26 | 978 | 984 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-3p | 60 | 67 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-342 | 1108 | 1115 | 1A,m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-363 | 545 | 551 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-369-3p | 745 | 752 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 746 | 752 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-377 | 1107 | 1113 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-450 | 989 | 995 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-495 | 202 | 208 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-500 | 544 | 550 | 1A | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-543 | 466 | 472 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-96 | 733 | 739 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.